New study. We compared the immune response of XBB and JN.1 in human infections to evaluate the necessity for #SARSCOV2 vaccine updates
Results:
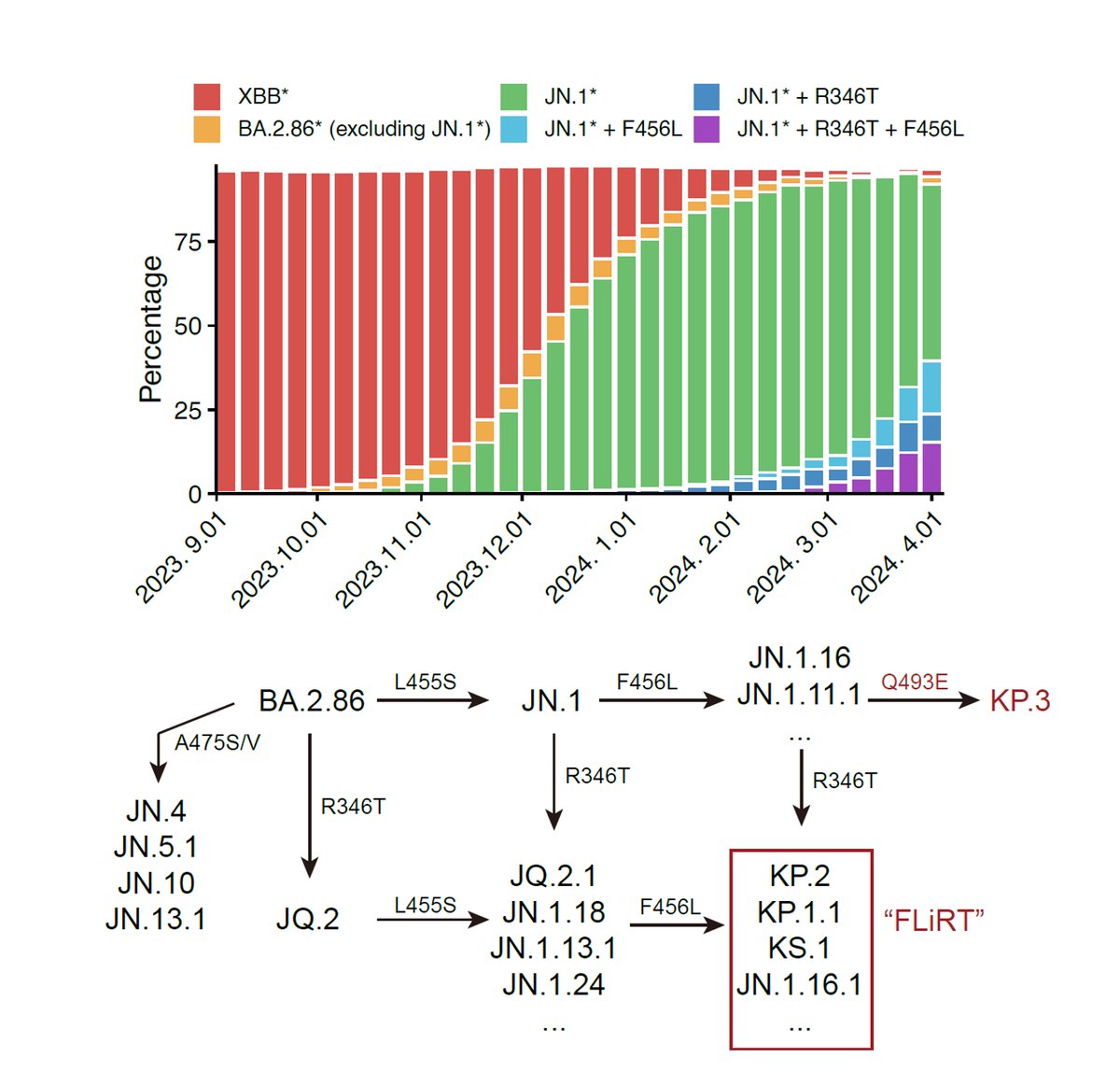
JN.1 exposure induces higher neutralization against emerging mutants, including FLiRT (JN.1+346T+456L) and KP.3
biorxiv.org/content/10.110…

Results:
JN.1 exposure induces higher neutralization against emerging mutants, including FLiRT (JN.1+346T+456L) and KP.3
biorxiv.org/content/10.110…

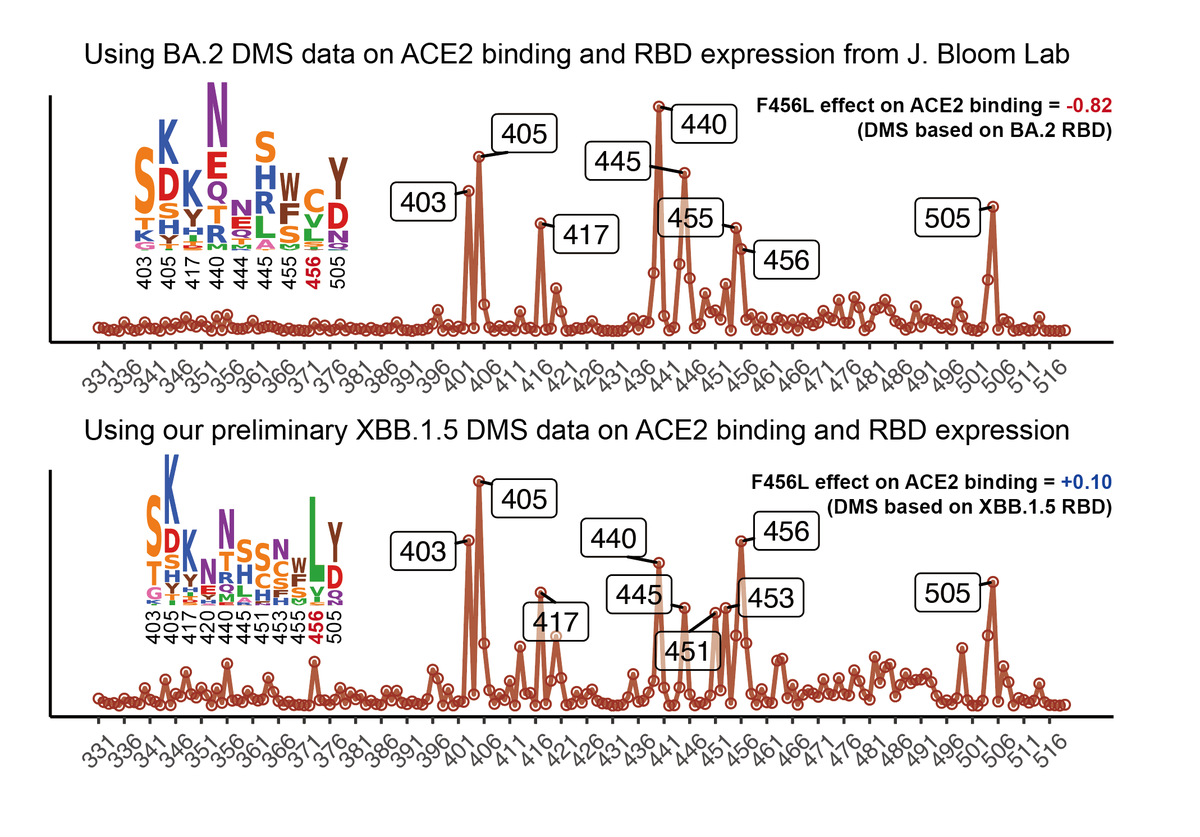
Since JN.1 lineages have replaced XBB lineages and JN.1 subvariants are continuously gaining immune-evasive mutations, such as R346T, F456L, R346T+F456L (FLiRT), and F456L+Q493E (KP.3), it's time to evaluate whether we need to switch SARS-CoV-2 vaccine antigen to JN.1.
(2/7)
(2/7)
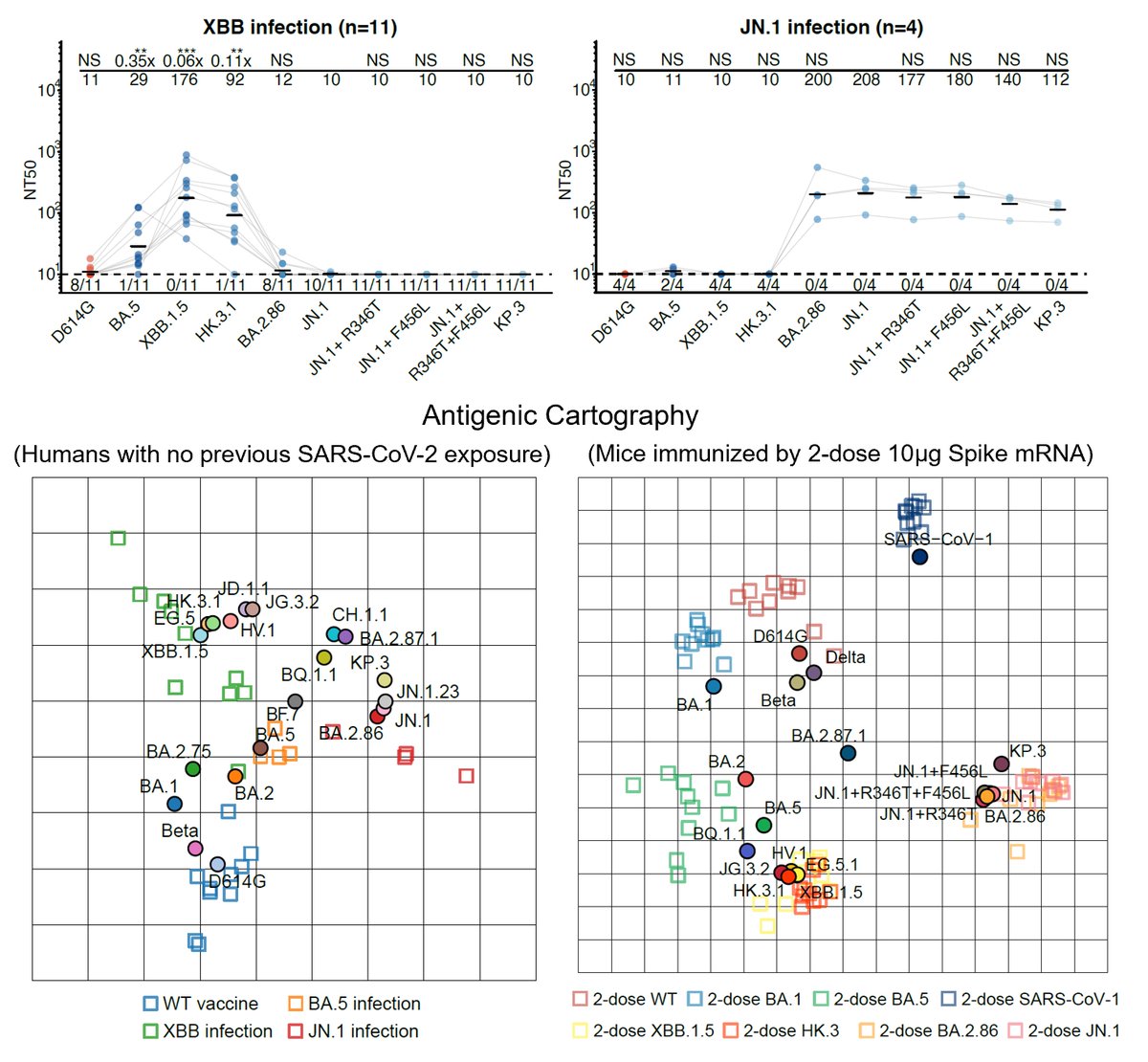
We first compared the antibody response of XBB and JN.1 infection in SARS-CoV-2 naive individuals (people who weren't vaccinated and haven't been infected). Similar to naive mice, we found that XBB and JN.1 lineages are also antigenic distinct in naive humans.
(3/7)
(3/7)
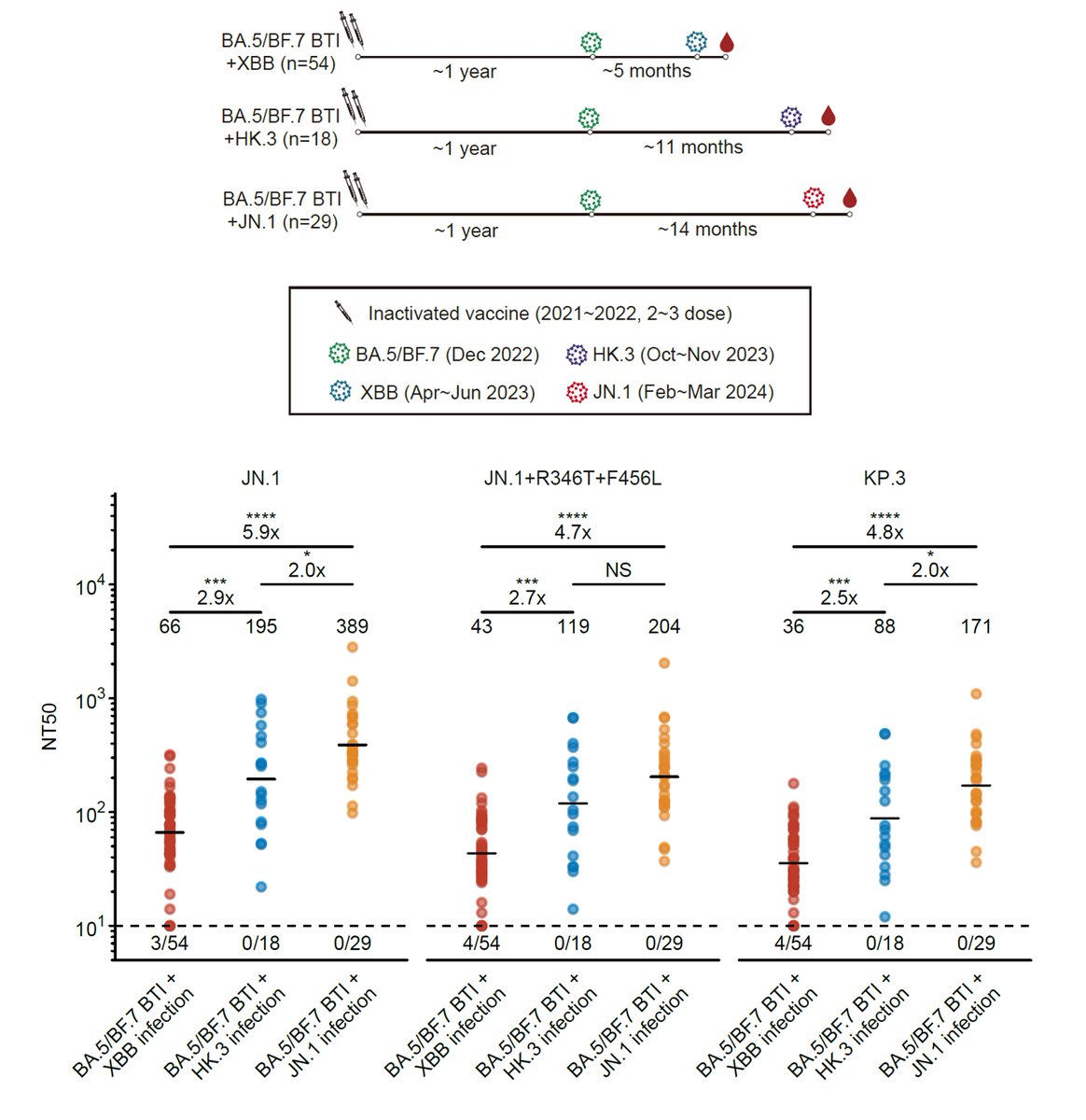
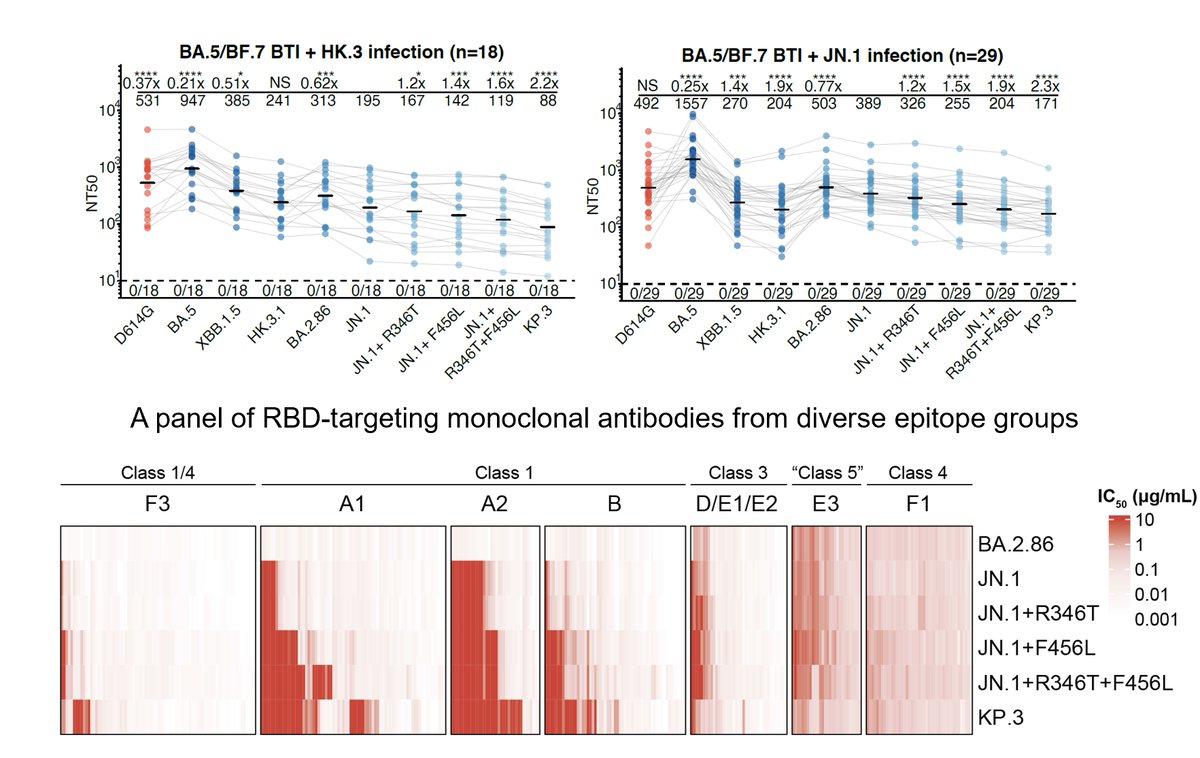
We then compared the immunogenicity of XBB and JN.1 infections in those who had been vaccinated and exposed to Omicron before (Major population). As expected, JN.1 exposure clearly induces higher neutralization titers against emerging mutants, such as FLiRT and KP.3.
(4/7)
(4/7)
KP.3 (JN.1+F456L+Q493E) is the most immune evasive variant we found and is also the fastest-growing JN.1 sublineage. The additional F456L and Q493E mutation allows KP.3 to evade a substantial proportion of JN.1-effective mAbs, especially Class 1 antibodies.
(5/7)
(5/7)
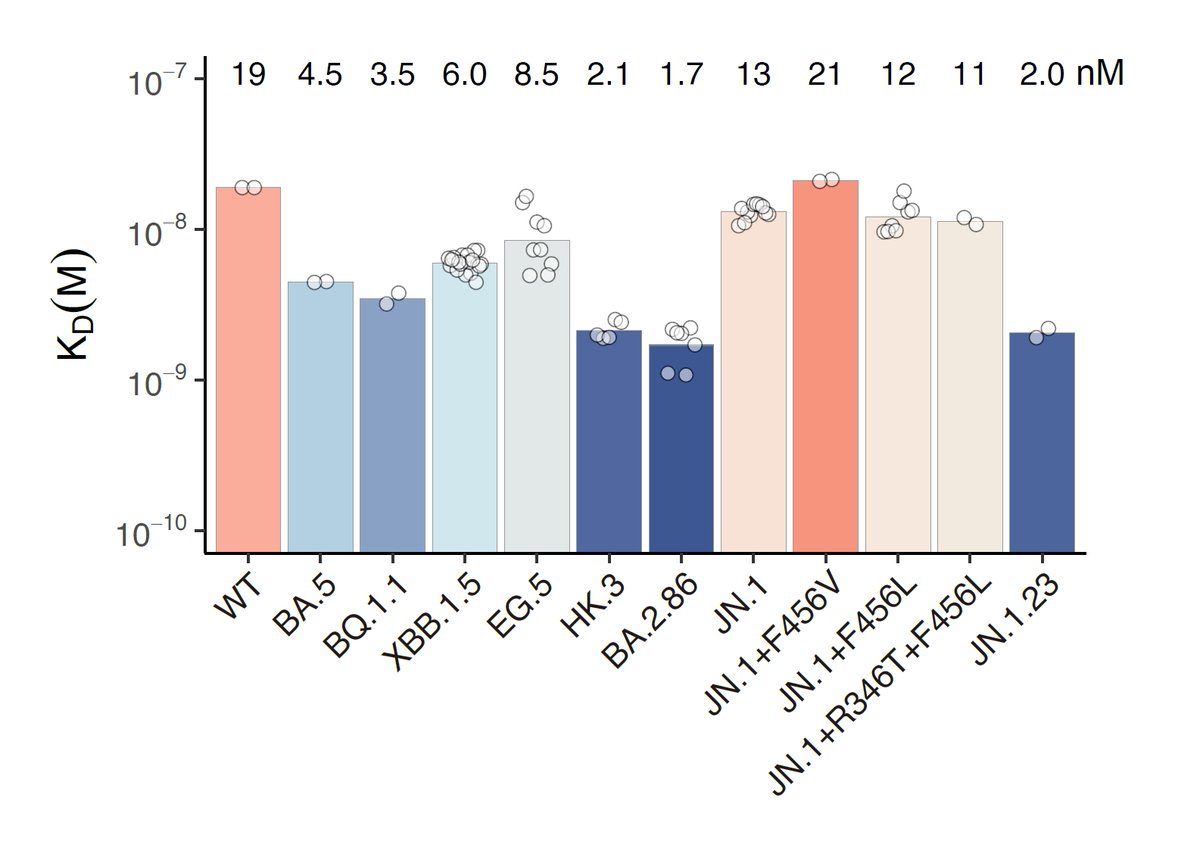
Additionally, we found that JN.1+F456L and FLiRT showed equal ACE2 binding affinity compared to JN.1, and JN.1.23, which carries a Y453F mutation, exhibits greatly enhanced ACE2 binding. Acquisition of more immune-evasive mutations should be closely monitored for JN.1.23.
(6/7)
(6/7)
We have also isolated and expressed mAbs from these cohorts to compare the humoral immune response landscape at the monoclonal antibody level of XBB and JN.1 exposure. Subsequent studies regarding the mAbs' epitope distribution and escaping mutations will soon be updated.
(7/7)
(7/7)
Current results do underscore the challenge posed by the continuously evolving SARS-CoV-2 JN.1 lineages and support the consideration of switching the focus of future SARS-CoV-2 vaccine updates to the JN.1 lineage.
• • •
Missing some Tweet in this thread? You can try to
force a refresh