KP.3 (w/the rare Q493E) has been my pick since I first noticed it emerging from numerous travel seqs from India. F456L & R346T are the typical stepwise immune-evasion mutations that, as @shay_fleishon noted, very likely impose a fitness cost. Q493E may be different. 1/
https://twitter.com/JosetteSchoenma/status/1785353182144610773
Q493E involves the rarest of all nucleotide mutations, C->G, and occurs at a key residue that we've seen very little action from of late. 493 mutations, however, are common in the Cryptics, usually Q493K I believe. (@SolidEvidence can correct me if I'm wrong on that). 2/8 
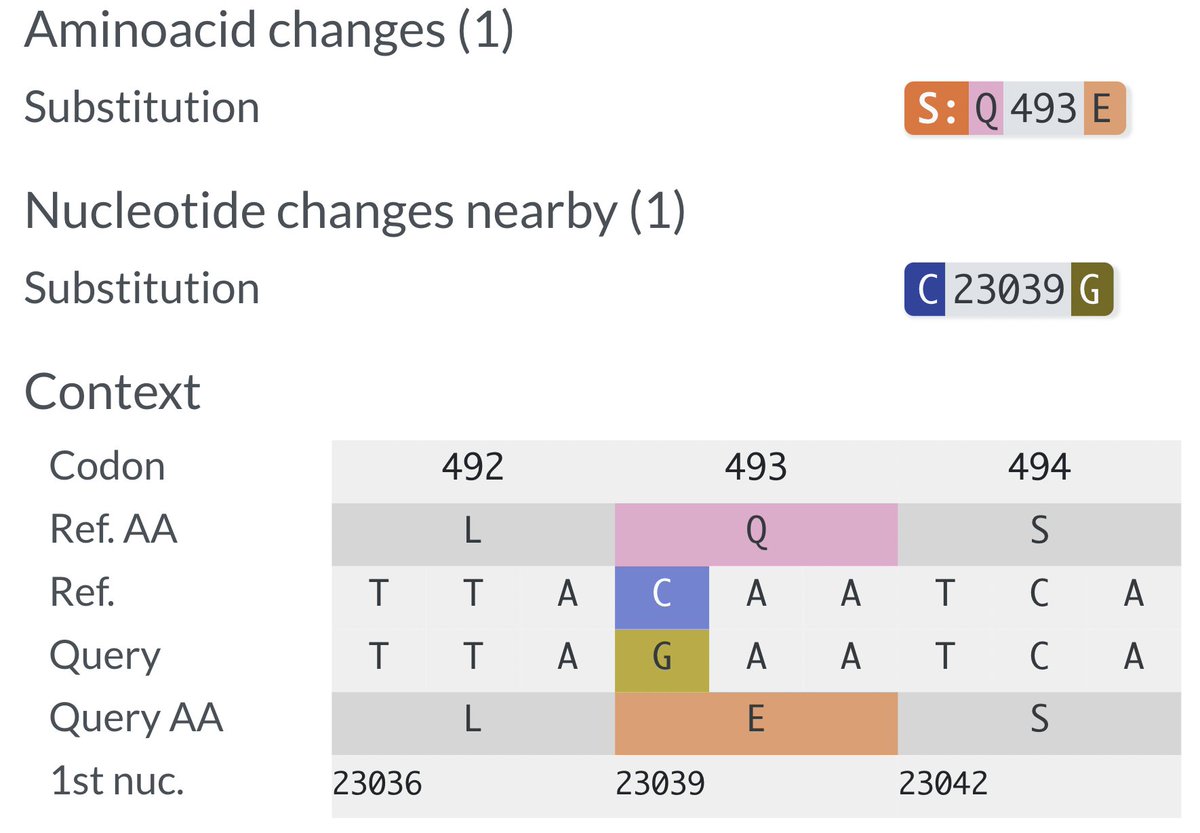
493 is also one of the few residues where mutations—on BA.1/BA.2 backgrounds—can confer large increases in ACE2 affinity—see @jbloom_lab data below. The 2-nuc Q493A & Q493V appeared in a handful of remarkable chronic-infection seqs, for example. 3/8 
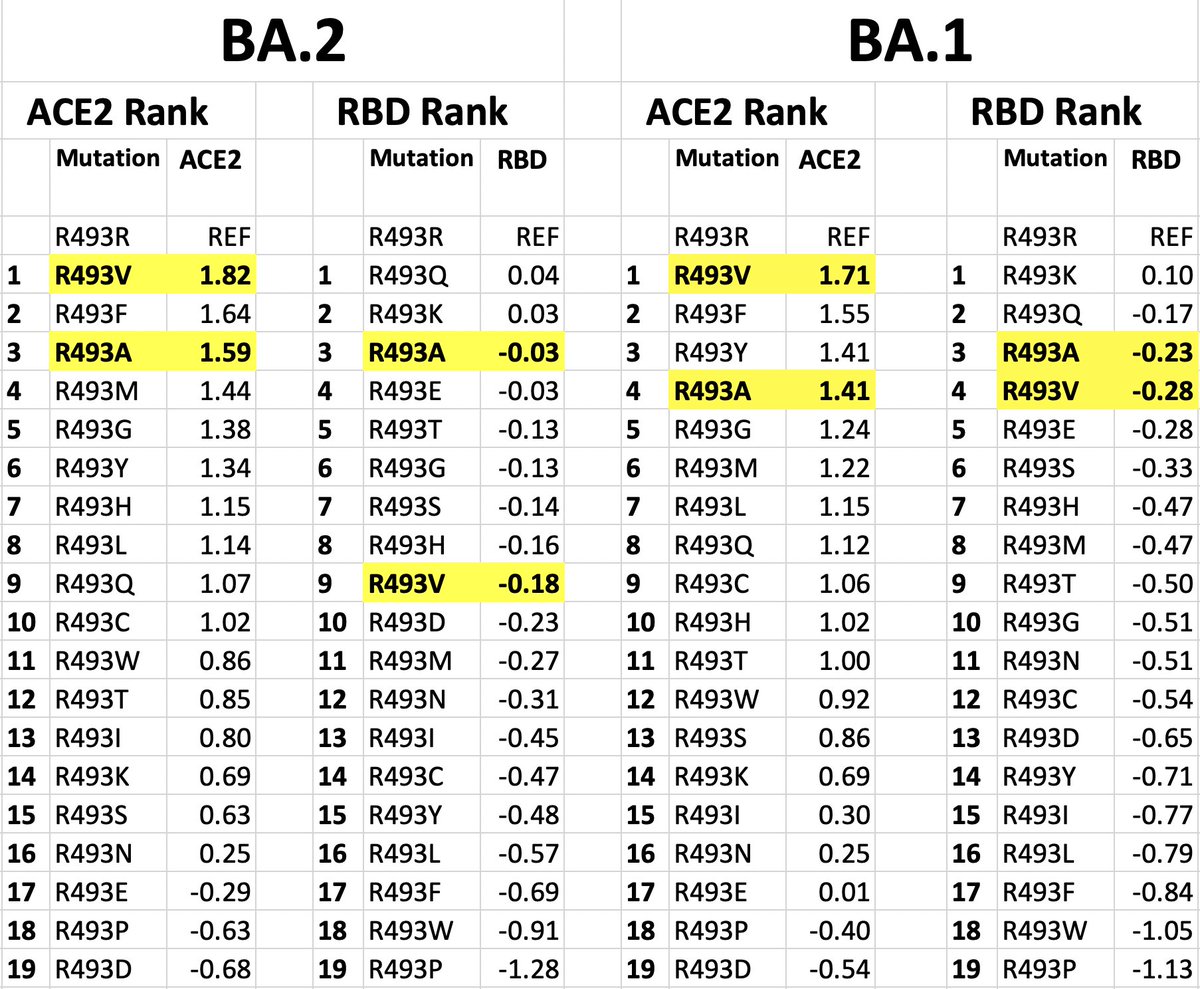
Q493E marks another major change in recent trends: Until now, the RBM (S:438-506) has become more & more basic (i.e. positively charged)—possibly to increase binding to co-receptor heparan sulfate, which @yunlong_cao showed is esp important for BA.2.86 4/
https://twitter.com/LongDesertTrain/status/1766259218808537351
With its V445H, N481K, & A484K mutations (slightly tempered by N450D), BA.2.86 represented the pinnacle of this basic RBM trend. Q493E, however, reverses this trend, making the RBM more acidic (negatively charged). Furthermore… 5/8
…Q493E is in close proximity to a lot of key spike AA residues, including 346 (R346T in FLiRT variants), 448-456, and the 483-484 region where BA.2.86 has ∆V483 & A484K.
h/t to @OliasDave for his spike AA proximity tool, which the data 👇 is from. 6/8 ukcovid.xyz/proximity.php?…

h/t to @OliasDave for his spike AA proximity tool, which the data 👇 is from. 6/8 ukcovid.xyz/proximity.php?…
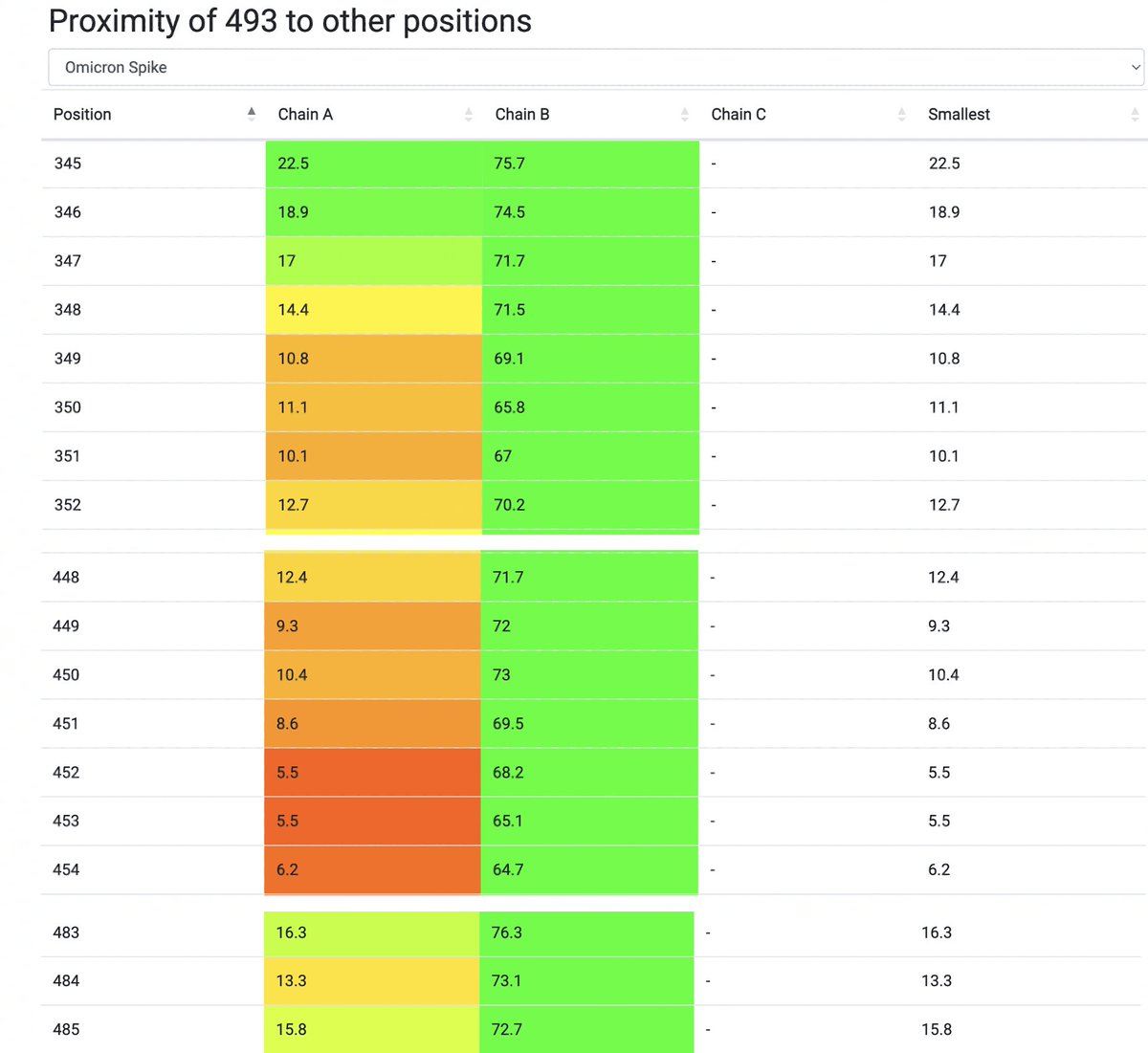
For reasons I've never understood, R346X mutations have never worked well with N450D. FLiRT variants have R346T + N450D, & right now, R346T's Ab-evasion powers give it a growth advantage. But I suspect the R346T-N450D combo still significantly weakens the virus. 7/8
KP.3's Q493E seems to put it on a clearly different path than the other converging JN variants. Baseline KP.3 is already outgrowing all the FLiRT variants. I suspect it has more room to tinker and improve & will continue to outgrow them in the coming weeks/months. 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh