Phylogenetic data support mammal-to-mammal transmission of H5N1 & Adaptation
Massive outbreak of Influenza A H5N1 in elephant seals at Peninsula Valdes, Argentina: increased evidence for mammal-to-mammal transmission
biorxiv.org/content/10.110…
Massive outbreak of Influenza A H5N1 in elephant seals at Peninsula Valdes, Argentina: increased evidence for mammal-to-mammal transmission
biorxiv.org/content/10.110…
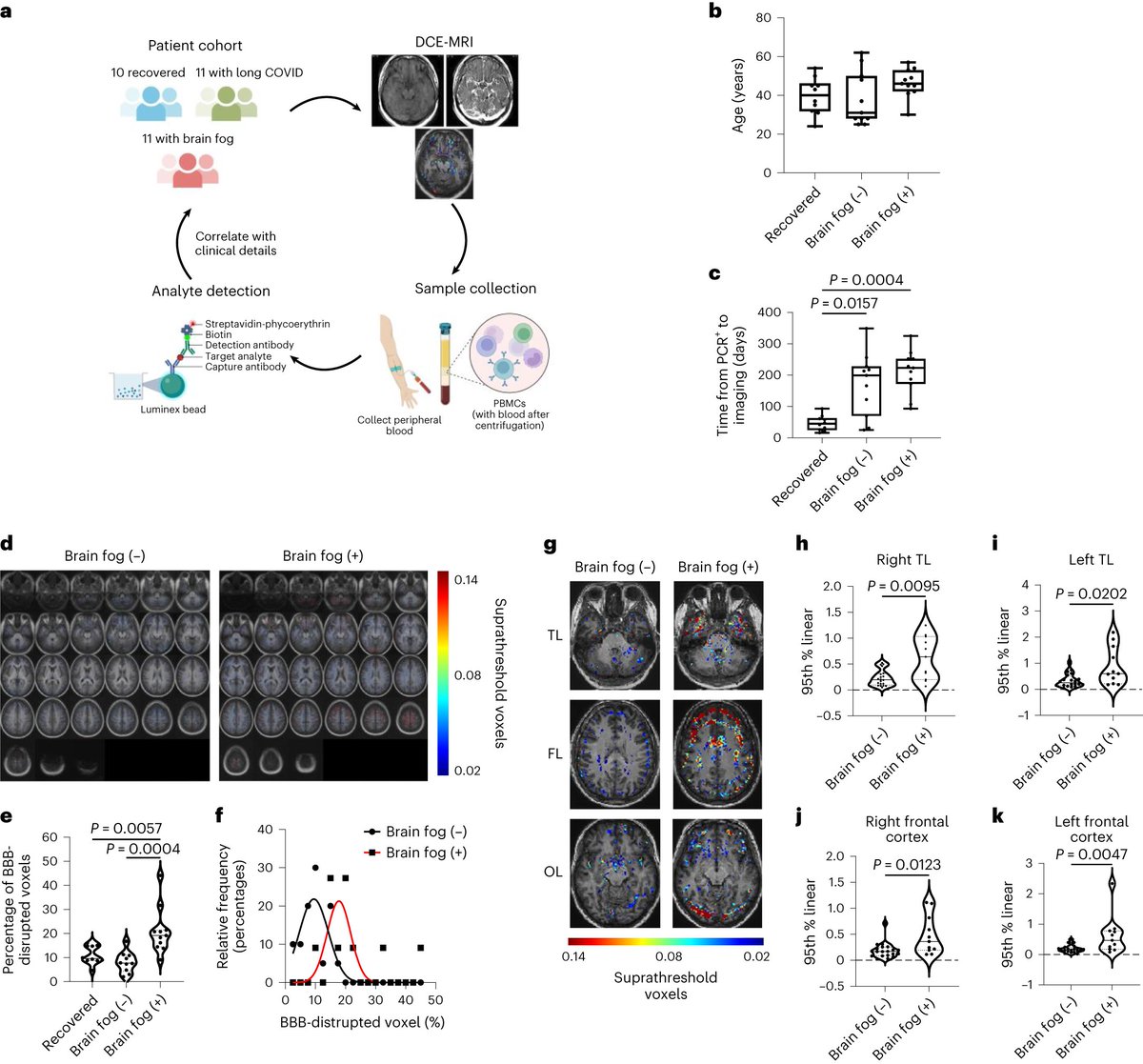
SNPs specific to the marine mammal clade:
* Q591K and D701N in PB2
* L548F in PB1
* A20T, M86I, and M548I in PA
* R21Q and I226T in NS1
* Q591K and D701N in PB2
* L548F in PB1
* A20T, M86I, and M548I in PA
* R21Q and I226T in NS1
"We believe that the high mortality rate in elephant seal pups is also consistent with mammal-to-mammal transmission, as pups are toothless and nurtured exclusively through nursing from their mothers."
"Some newborns may have been infected before birth, as transplacental transmission of H5N1 HPAI viruses has been reported in humans and high virus loads were detected in aborted sea lion fetuses."
"Mammal-to-mammal transmission is also supported by our regional phylogenetic analysis, which identified a novel H5N1 2.3.4.4b clade with viruses that appear to be specific to marine mammals. This marine mammal clade comprises strains with mutations..."
"...that were not present in H5N1 2.3.4.4b viruses in birds (wild and domestic) from Peru, Chile, Argentina, Uruguay and Brazil, excepting occasional spillovers from marine mammals to coastal birds (terns and sanderling)."
They authors note that ecologically it is unlikely adults are getting H5N1 infections via food sources or interactions with birds. South American sea lions regularly visit multiple rookeries and haul-outs and likely spread H5N1 directly throughout Venezuela, Uruguay and Brazil.
Some of these mutations (such as Q591K and D701N in PB2) are associated with increased virulence, transmission, or adaptation to mammalian hosts and have been maintained since they first emerged in H5N1 HPAI viruses in marine mammals in Chile.
That the novel H5N1 marine mammal clade had an independent chain of virus transmission is evidenced by the slow rate of viral evolution and it's widespread distinct pathways of spread across host groups and geographical areas.
Phylogenetic tree of H5N1 HPAI (2.3.3.4b) viruses in South America. Time-scale MCC tree inferred for the concatenated genome sequences (~13kb) of 236 H5N1 influenza A viruses (clade 2.3.3.4b) collected in five South American countries (Argentina, Brazil, Chile, Peru, Uruguay) an Antarctica. Three spillover events into marine mammals, including the marine mammal clade, are labeled Branches are shaded by inferred host species and location (13 categories). Posterior probabilities provided for key nodes. Tip labels provided for all mammalian viruses and a selection of avian viruses. See page 28 of PDF.

Posterior distributions of evolutionary rates (substitutions per site per year) inferred for the complete virus genome (all positions) and for only the third nucleotide position for H5N1 (2.3.4.4b) in South America, partitioned into two host categories: marine mammal and wild bird/poultry.

Mutations defining the marine mammal clade of H5N1 HPAI (2.3.4.4b) viruses. Amino acid changes are listed for new mutations that arose in the marine mammal clade of the H5N1 HPAI (2.3.4.4b) viruses that are not observed in any other avian viruses included in this study from South America, mapped against the subsection of the MCC tree with the marine mammal clade Virus names and associated mutations are colored by country. Location/month of collection (in 2023) are listed for Argentina and Brazil. A question mark indicates that no sequence data is available at that position for that virus. H5 numbering is used for HA.

Chronology and hypothesized pathways of spread of H5N1 HPAI (2.3.4.4b) viruses in South America, 2022–2023. H5Nx HPAI detections (1-Sep-2022 to 1-Nov-2023) reported to the World Animal Health Information System (WAHIS/WOAH) are represented with orange circles (wild birds), green triangles (domestic birds) and blue squares (mammals). The location of the outbreak investigated in this study (Península Valdés) is highlighted in red. Arrows represent the timeline of hypothesized pathways of virus spread, as derived from the chronology of detections and our phylodynamic analysis. The pathways of virus spread and significant events of the avian and marine mammal clade viruses are represented in dark orange and dark blue, respectively. Dark yellow represents incidental avian hosts of marine mammal clade viruses (i.e. spillover). Note that virus spread pathways in this figure are intended as a conceptual model and are not geographically precise.

• • •
Missing some Tweet in this thread? You can try to
force a refresh