What if we could universally recombine, insert, delete, or invert any two pieces of DNA?
In back-to-back @Nature papers, we report the discovery of bridge RNAs and 3 atomic structures of the first natural RNA-guided recombinase - a new mechanism for programmable genome design
In back-to-back @Nature papers, we report the discovery of bridge RNAs and 3 atomic structures of the first natural RNA-guided recombinase - a new mechanism for programmable genome design
See our previous thread on preprint here:
Bridge RNA recombinases encode a mechanistically new class of guide RNAs
They are bispecific, unlike the CRISPR (2012) or RNA interference (1998) guide RNAs that specify only 1 target for a simple scissors cut

Bridge RNA recombinases encode a mechanistically new class of guide RNAs
They are bispecific, unlike the CRISPR (2012) or RNA interference (1998) guide RNAs that specify only 1 target for a simple scissors cut
https://x.com/pdhsu/status/1750316518133694734

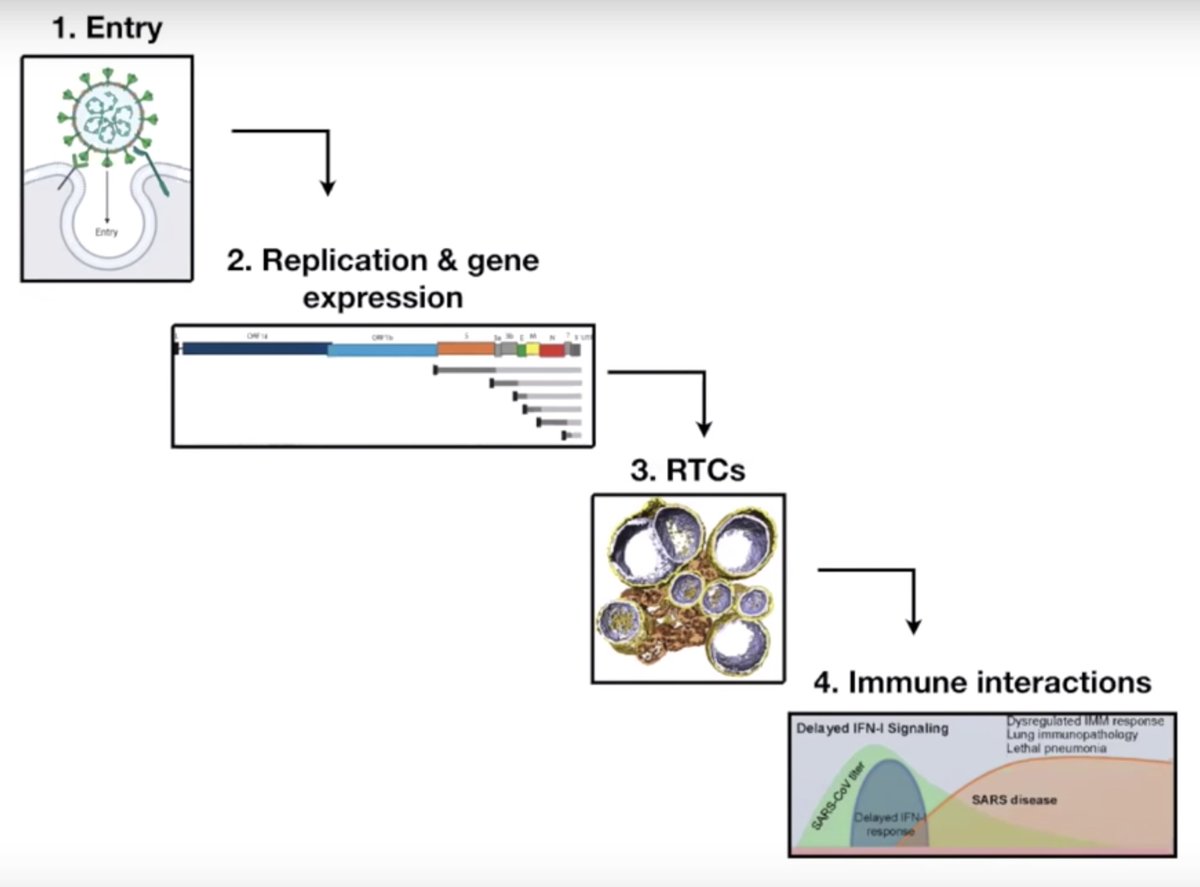
In new collaborative work with @hnisimasu, we reveal the IS110 bridge mechanism in 3 distinct stages
The bispecific bridge RNA recruits two dimers of the IS110 recombinase, forming an elegant tetramer to exchange and religate 4 distinct DNA strands in a single step!
The bispecific bridge RNA recruits two dimers of the IS110 recombinase, forming an elegant tetramer to exchange and religate 4 distinct DNA strands in a single step!
CRISPR creates DNA nicks/breaks and requires complex DNA repair pathways in the cell to make a genome edit (like an insertion or deletion) beyond a cut
Bridge recombination works in vitro, without relying on cellular DNA repair mechanisms. This could lead to safer genome edits
Bridge recombination works in vitro, without relying on cellular DNA repair mechanisms. This could lead to safer genome edits

Cre-Lox recombination has been a foundational tool for molecular biology and mouse genetics since its discovery in 1981. However, it is not programmable.
Scientists have spent decades trying to engineer and reprogram Cre to recognize DNA sites other than the LoxP sequence
Scientists have spent decades trying to engineer and reprogram Cre to recognize DNA sites other than the LoxP sequence

Unlike Cre, bridge recombination is fully programmable simply by changing the target binding loop (left) or donor binding loop (right)
These two loops can be modularly and separably reprogrammed
These two loops can be modularly and separably reprogrammed

Inspired by Cre engineering, we show that reprogrammed bridge RNAs can direct insertion, excision, or inversion of DNA
This is a universal mechanism that makes the desired DNA rearrangement end-to-end in a single step
This is a universal mechanism that makes the desired DNA rearrangement end-to-end in a single step

How does this enable large-scale genome design beyond editing individual bases or genes?
The experimental ability to universally rearrange/recombine DNA must be paired with the computational ability to design sequences at genome-scale
That's Evo, our AI foundation model with @BrianHie trained on genomes:
The experimental ability to universally rearrange/recombine DNA must be paired with the computational ability to design sequences at genome-scale
That's Evo, our AI foundation model with @BrianHie trained on genomes:
https://x.com/pdhsu/status/1762512557565456825
Together, I think these studies open up entirely new directions and we are excited to explore them through many collaborations
The bridge discovery was co-led by the amazing duo of @mgdurrant and @ntperry13 with important contributions from @SKonermann, @jjwp1011, @Adi_R_Jangid, @hnisimasu, Masa, @AprilPawluk, @JSAthukoralage, @johnmcspeedy
The structural mechanism studies were led by @hnisimasu and Masahiro Hiraizumi. Hiroshi and I first started working together over 10 years ago, on the first atomic structure of Cas9 bound to guide RNA and target DNA ()
One of the great pleasures of a life in science is long-term collaboration with wonderful friends and colleagues, and I'm grateful to continue teaming up with Hiroshi and also the brilliant @SKonermann (whom I've worked with for nearly 15 years now across dozens of papers and, of course, starting @arcinstitute)cell.com/fulltext/S0092…
The bridge discovery was co-led by the amazing duo of @mgdurrant and @ntperry13 with important contributions from @SKonermann, @jjwp1011, @Adi_R_Jangid, @hnisimasu, Masa, @AprilPawluk, @JSAthukoralage, @johnmcspeedy
The structural mechanism studies were led by @hnisimasu and Masahiro Hiraizumi. Hiroshi and I first started working together over 10 years ago, on the first atomic structure of Cas9 bound to guide RNA and target DNA ()
One of the great pleasures of a life in science is long-term collaboration with wonderful friends and colleagues, and I'm grateful to continue teaming up with Hiroshi and also the brilliant @SKonermann (whom I've worked with for nearly 15 years now across dozens of papers and, of course, starting @arcinstitute)cell.com/fulltext/S0092…

This was a 2+ year detective story uniquely enabled by the @arcinstitute model, blending Arc's technical staff, PhD students from our partner universities, and hybrid computational & experimental science.
Matt, Nick, and the team have moved mountains to bring you this tour-de-force work, which combines ideas, scholarship, and methods across the fields of computational biology, genetics, biochemistry, molecular biology, microbiology, structural biology, and bioengineering - all in a single project.
Matt, Nick, and the team have moved mountains to bring you this tour-de-force work, which combines ideas, scholarship, and methods across the fields of computational biology, genetics, biochemistry, molecular biology, microbiology, structural biology, and bioengineering - all in a single project.

Moving forward, I'm particularly interested in programming biology across multiple lengthscales, including tissues and physiology.
We're hiring across ML and experimental positions for folks interested in combining biology and AI. Come join us at @arcinstitute!
Here are the two papers:
nature.com/articles/s4158…
nature.com/articles/s4158…
We're hiring across ML and experimental positions for folks interested in combining biology and AI. Come join us at @arcinstitute!
Here are the two papers:
nature.com/articles/s4158…
nature.com/articles/s4158…

Thanks also to @connorjtou and @BKleinstiver for this great News and Views in @Nature on our two studies!
nature.com/articles/d4158…
nature.com/articles/d4158…
• • •
Missing some Tweet in this thread? You can try to
force a refresh