@ArcInstitute co-founder, @BerkeleyBioE professor, @ThriveCapital investor | biology and ML research | 🇨🇦 prev @harvard @broadinstitute, Fast Grants
2 subscribers
How to get URL link on X (Twitter) App


 Evo 2 is trained on 9.3T tokens of DNA with single-base resolution at 1M token context length 🥳
Evo 2 is trained on 9.3T tokens of DNA with single-base resolution at 1M token context length 🥳

 COMBINE revealed diverse synergistic 🤝 and antagonistic 🤬 interactions between epigenetic protein domains, which we were able to study by calculating "synergy scores" for each individual pair and broader domain class (HDACs shown below)
COMBINE revealed diverse synergistic 🤝 and antagonistic 🤬 interactions between epigenetic protein domains, which we were able to study by calculating "synergy scores" for each individual pair and broader domain class (HDACs shown below) 

https://x.com/pdhsu/status/1750316518133694734



 This was a remarkable detective story 🕵️that starts with transposons. These mobile genetic elements (MGEs) jump around genomes 🧬 and play a central and ancient role in evolution and speciation. IS110 is a poorly understood family of MGEs with unusually long noncoding sequences!
This was a remarkable detective story 🕵️that starts with transposons. These mobile genetic elements (MGEs) jump around genomes 🧬 and play a central and ancient role in evolution and speciation. IS110 is a poorly understood family of MGEs with unusually long noncoding sequences! 

 saap ver (“damn good” in Thai) is the best Thai place in the city. the ambiance is boisterous while the food comes correct: an “asian spicy” order puts you straight into a fragrant hot ones episode
saap ver (“damn good” in Thai) is the best Thai place in the city. the ambiance is boisterous while the food comes correct: an “asian spicy” order puts you straight into a fragrant hot ones episode


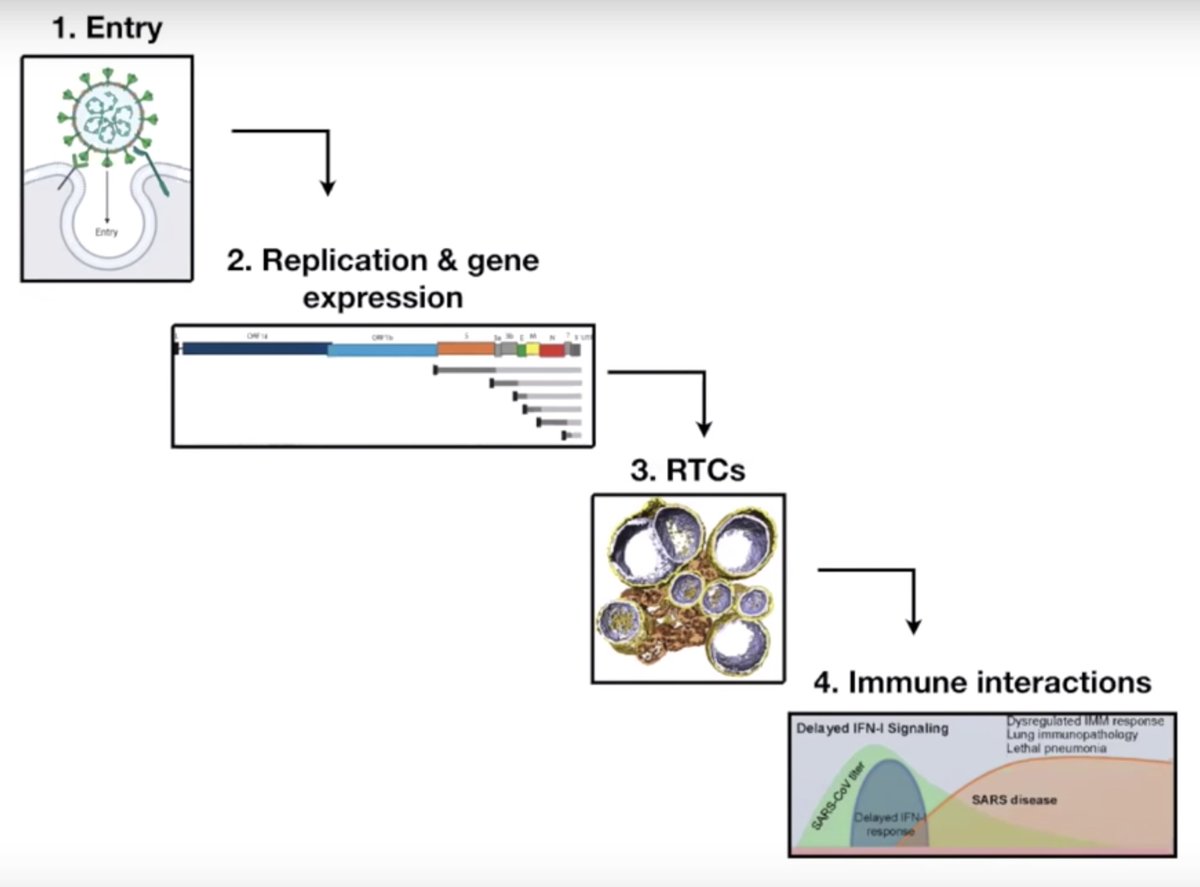
 An exonuclease (ExoN) is encoded in coronaviruses to enable their super large (30 kb) genomes. ExoN protects CoVs from inhibition by nucleoside analogs (e.g. Remdesivir @GileadSciences), suggesting a combo treatment w/ Remdesivir and an ExoN inhibitor could be effective
An exonuclease (ExoN) is encoded in coronaviruses to enable their super large (30 kb) genomes. ExoN protects CoVs from inhibition by nucleoside analogs (e.g. Remdesivir @GileadSciences), suggesting a combo treatment w/ Remdesivir and an ExoN inhibitor could be effective 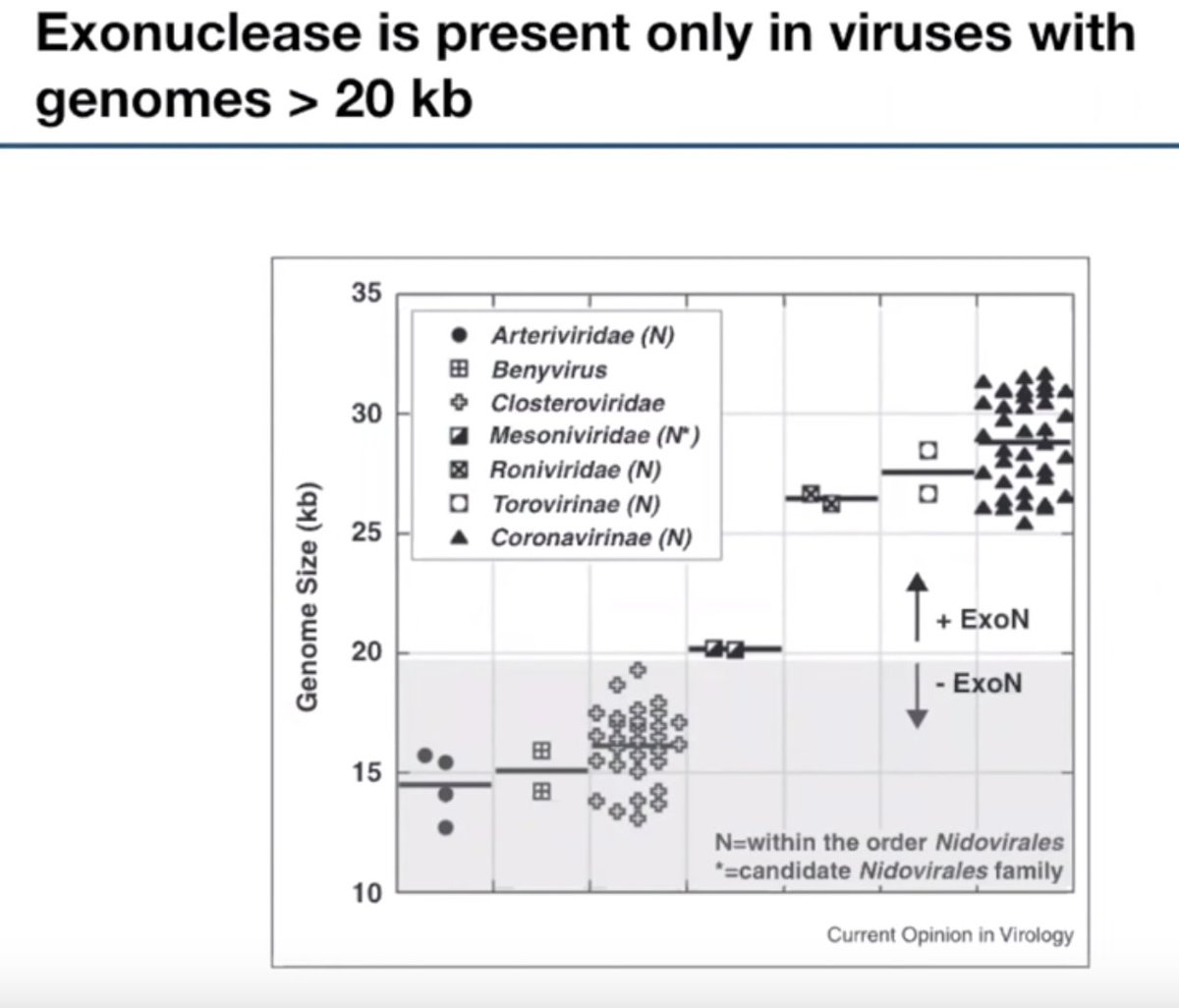
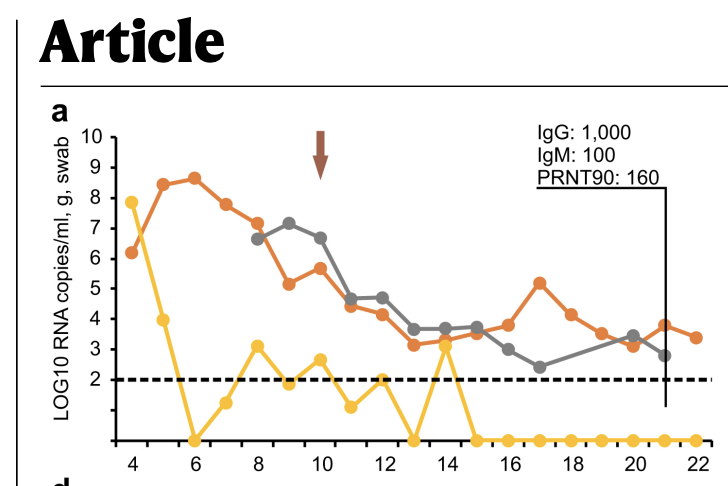
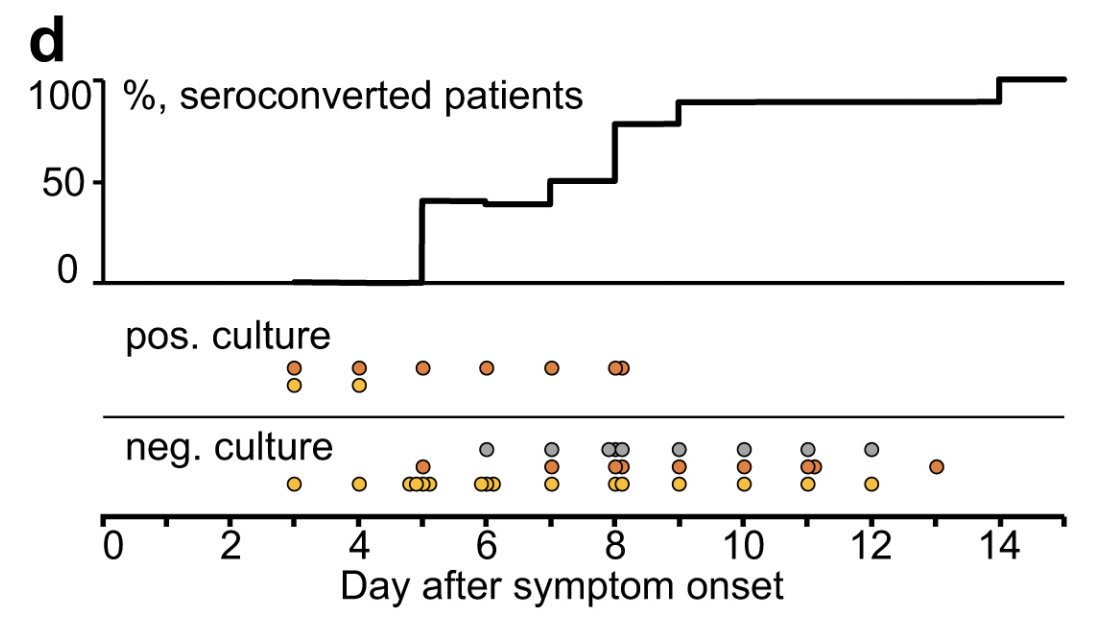
 Link to study, sorry: nature.com/articles/s4158…
Link to study, sorry: nature.com/articles/s4158…