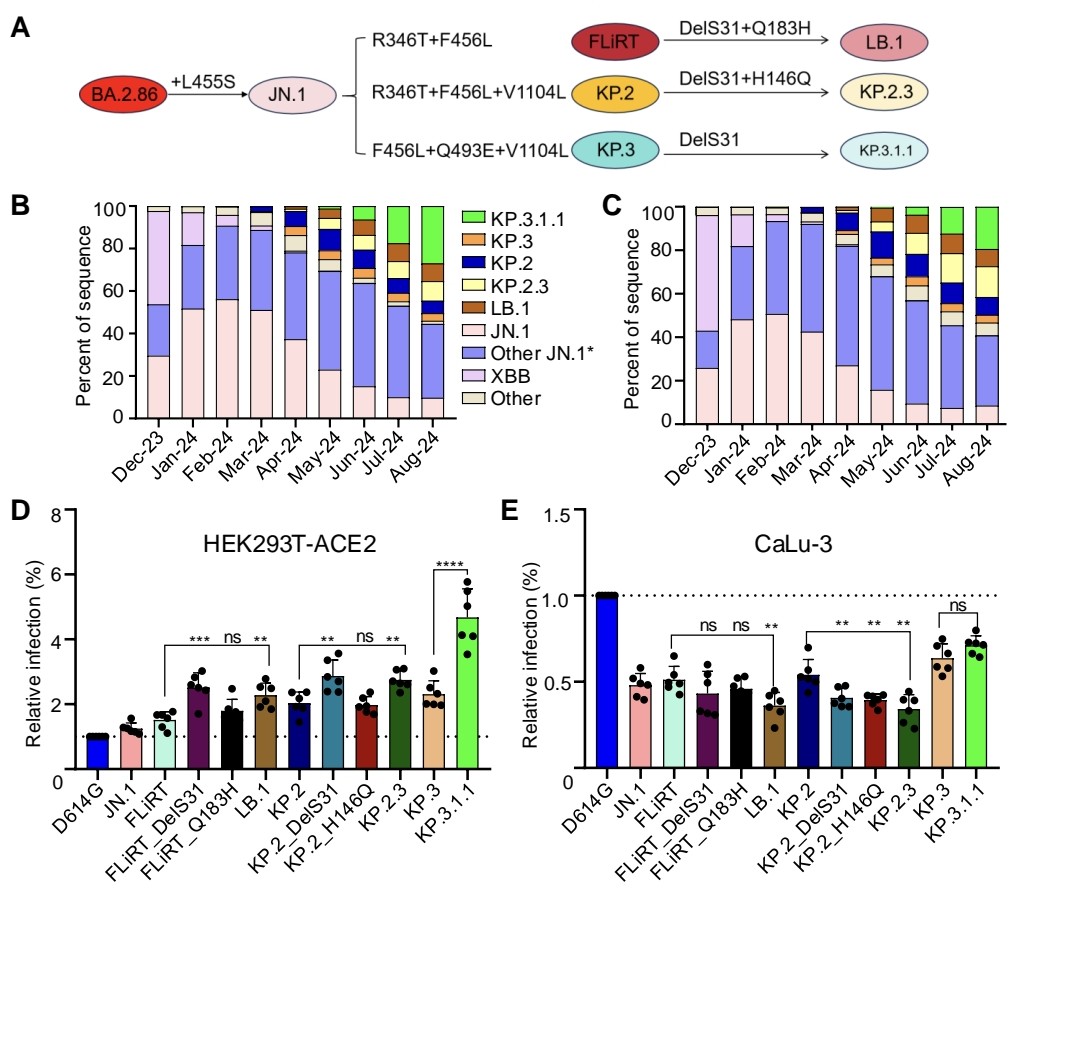
COVID PATIENTS EXHALE as many as 1,000 COPIES of the VIRUS PER MINUTE during the FIRST EIGHT DAYS of infection !!!
Article including a very interesting video :
news.northwestern.edu/stories/2023/0…

Article including a very interesting video :
news.northwestern.edu/stories/2023/0…

2) Study :
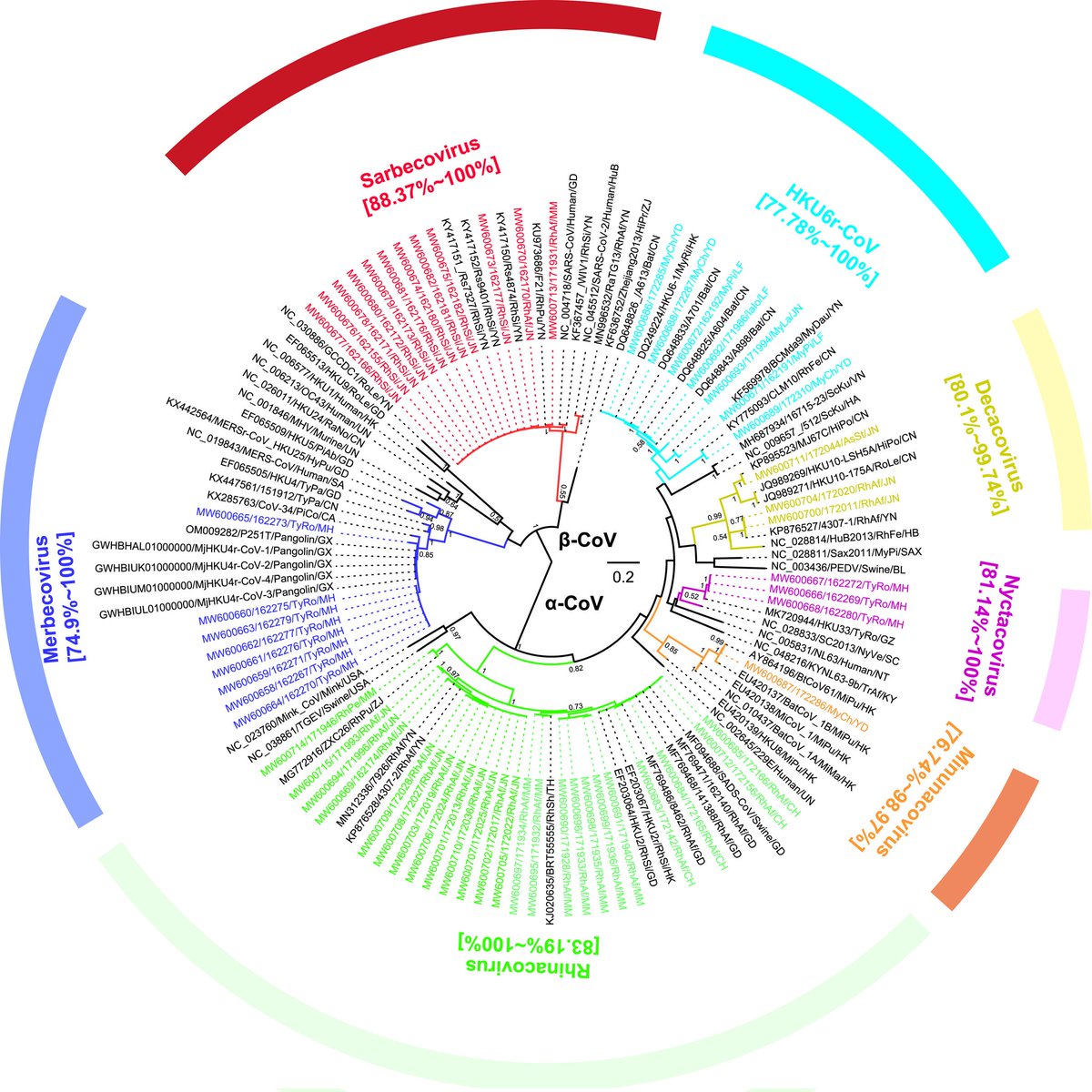
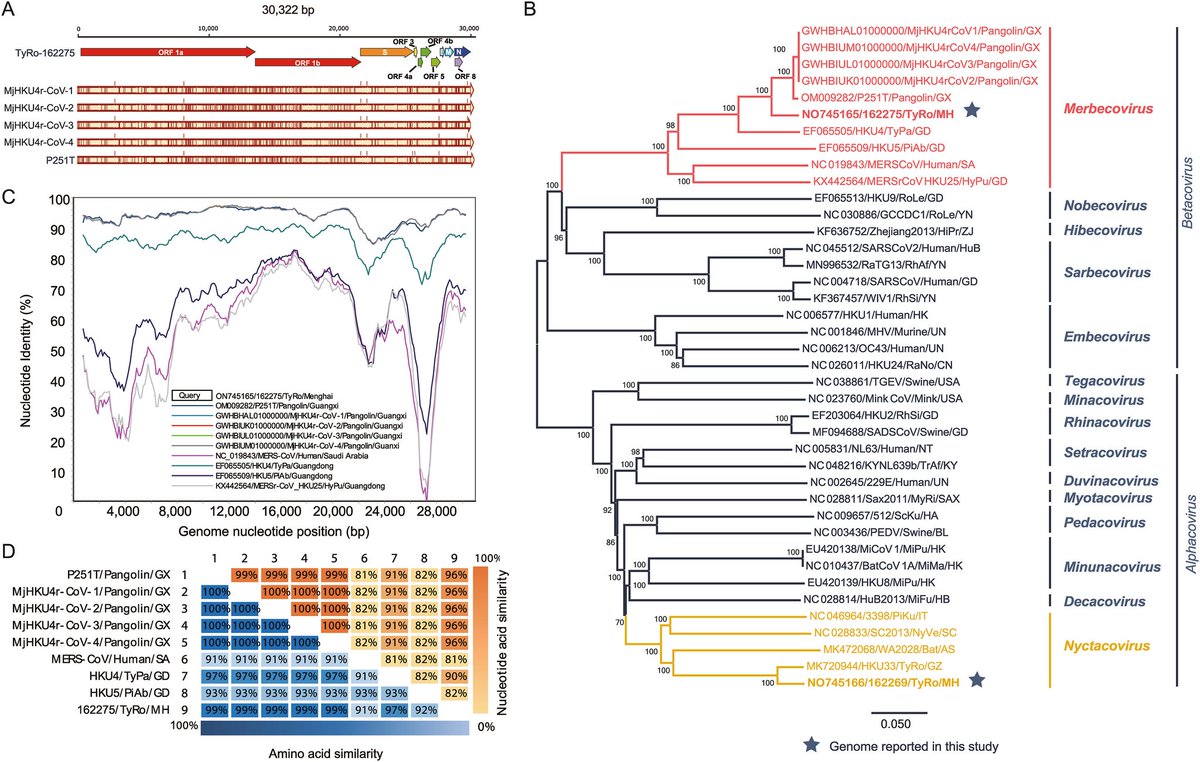
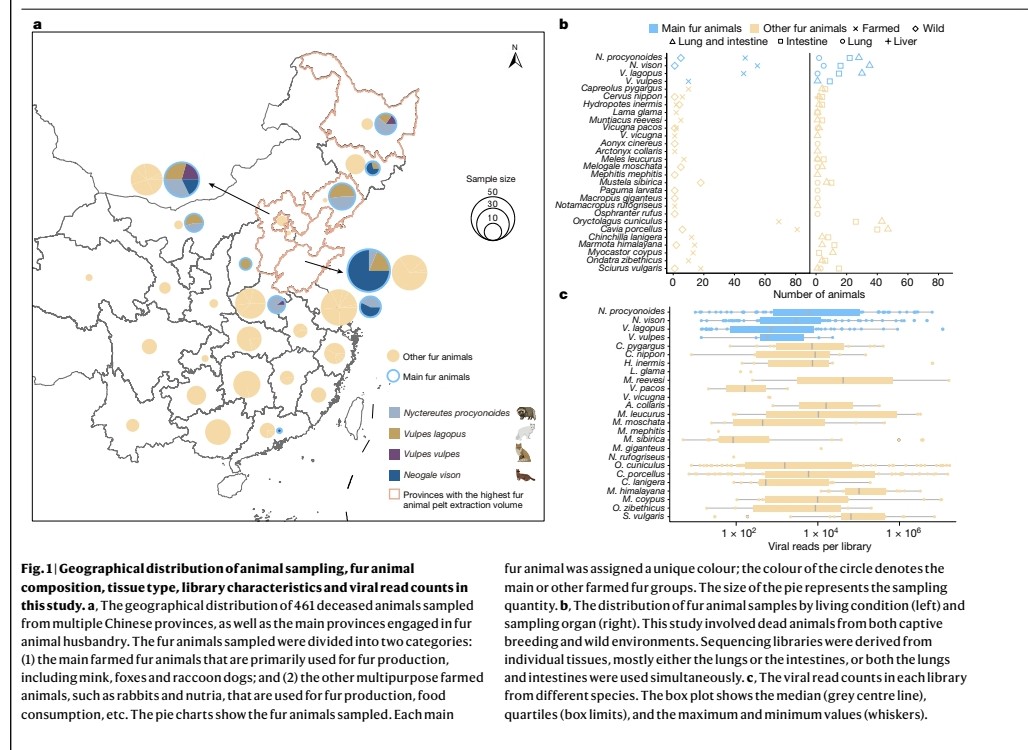
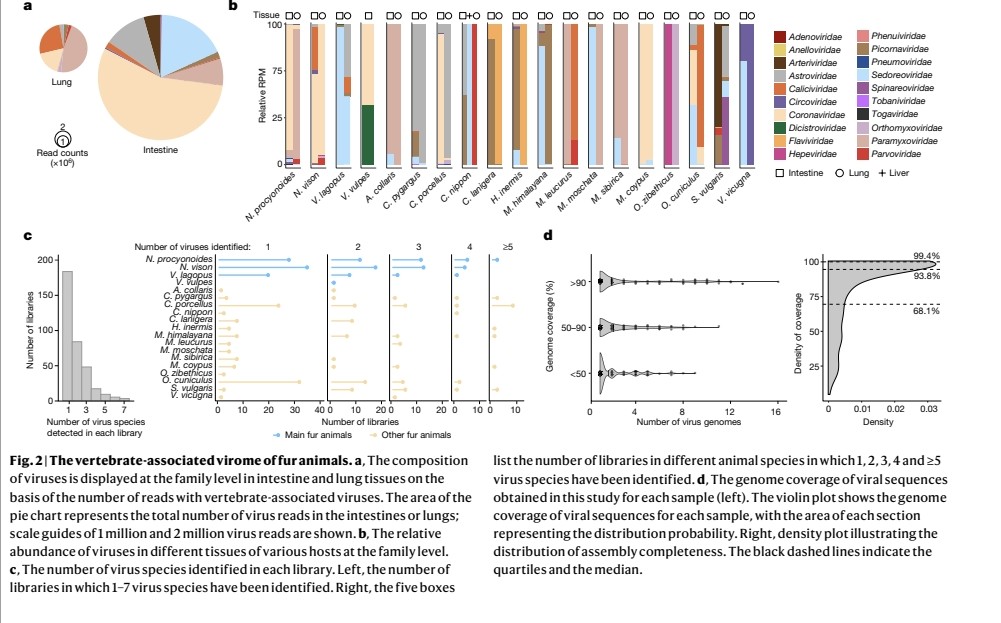
"Quantity of SARS-CoV-2 RNA copies exhaled per minute during natural breathing over the course of COVID-19 infection"
medrxiv.org/content/10.110…

"Quantity of SARS-CoV-2 RNA copies exhaled per minute during natural breathing over the course of COVID-19 infection"
medrxiv.org/content/10.110…
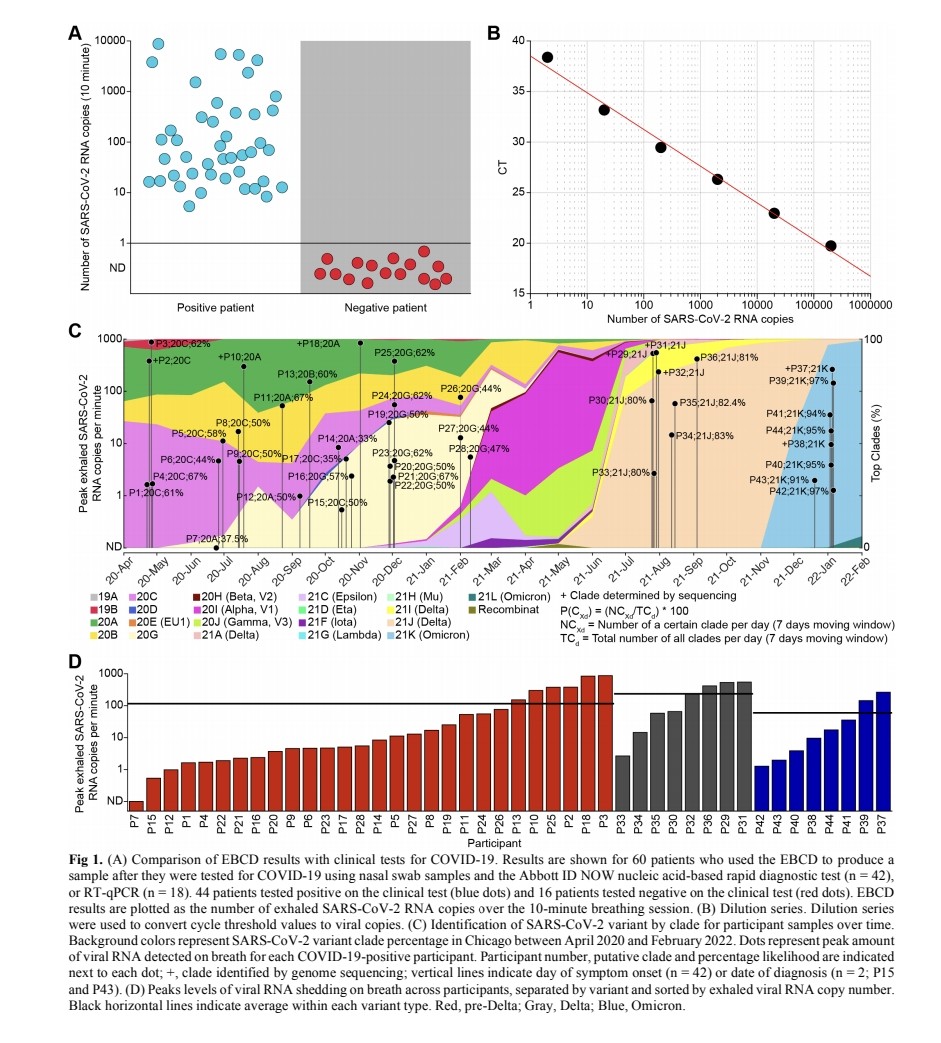
3) The study used a new portable device to collect exhaled breath samples from COVID-19 patients and measure the amount of SARS-CoV-2 RNA being expelled during natural breathing.
4) The main findings are:
- Infected patients exhaled an average of 80 viral RNA copies per minute during the first 8 days of infection, with high variability including peaks over 800 copies/minute.
- Infected patients exhaled an average of 80 viral RNA copies per minute during the first 8 days of infection, with high variability including peaks over 800 copies/minute.
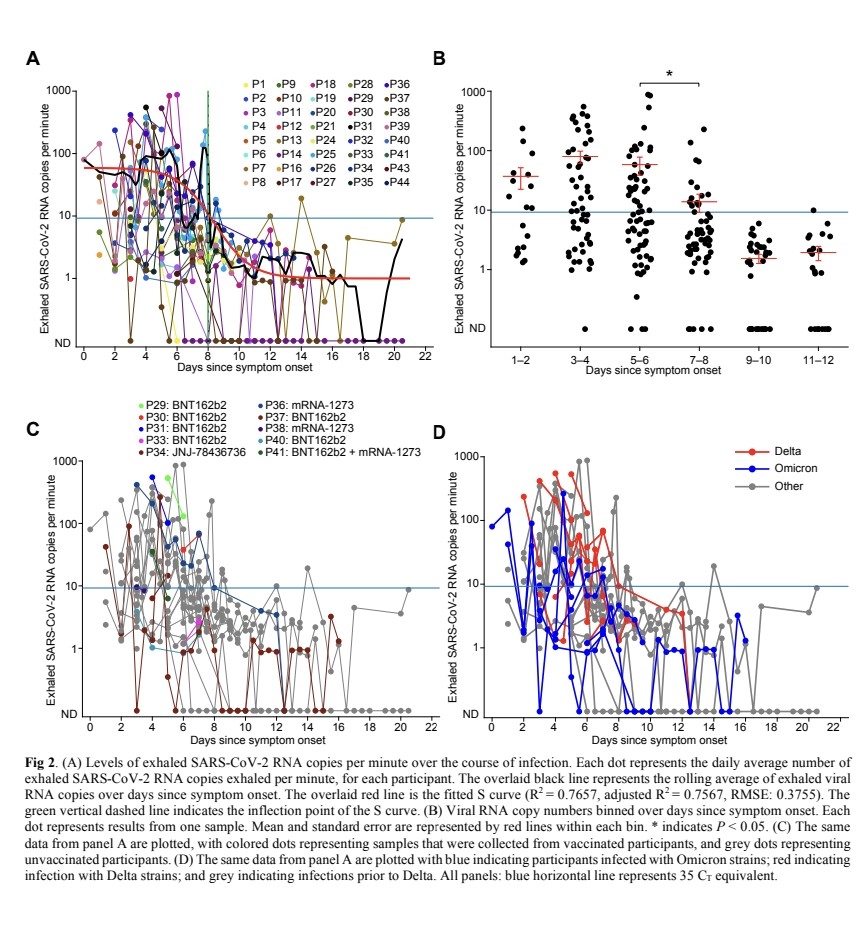
5) Viral RNA levels on breath did not significantly decrease until around day 8 from symptom onset, then dropped steeply to near the limit of detection.
Exhaled viral loads increased with self-reported symptom severity, though with high individual variation.
Exhaled viral loads increased with self-reported symptom severity, though with high individual variation.
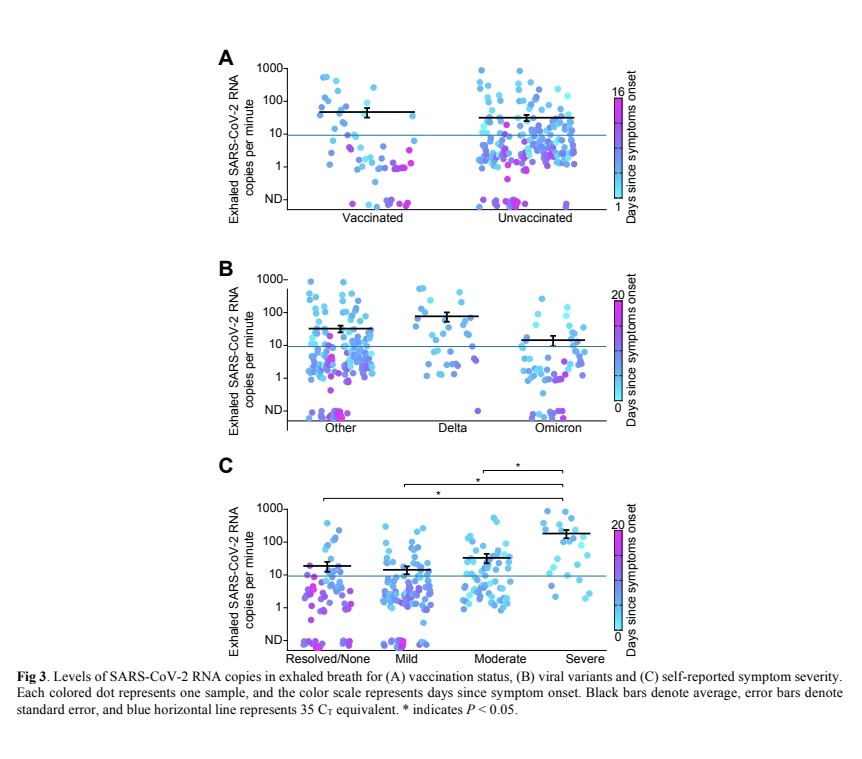
6) Exhaled viral loads did not differ based on vaccination status, viral variant, age, sex, or presence of co-morbidities.
The study provides important data on the dynamics of SARS-CoV-2 shedding in exhaled breath over the course of infection ...
The study provides important data on the dynamics of SARS-CoV-2 shedding in exhaled breath over the course of infection ...
7) ...which is critical for understanding transmission risk. The new portable collection device enabled frequent sampling to capture this temporal profile.
Thanks for reading 🙏
Thanks for reading 🙏
• • •
Missing some Tweet in this thread? You can try to
force a refresh