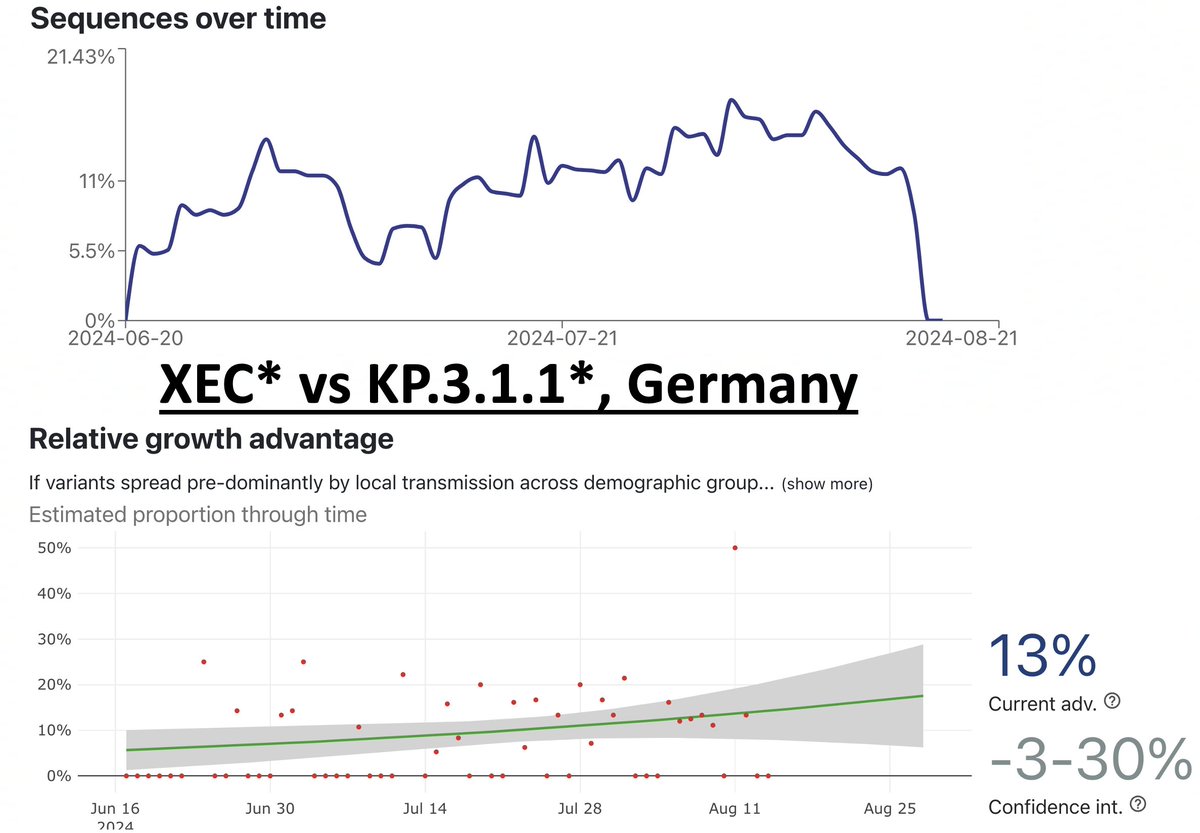
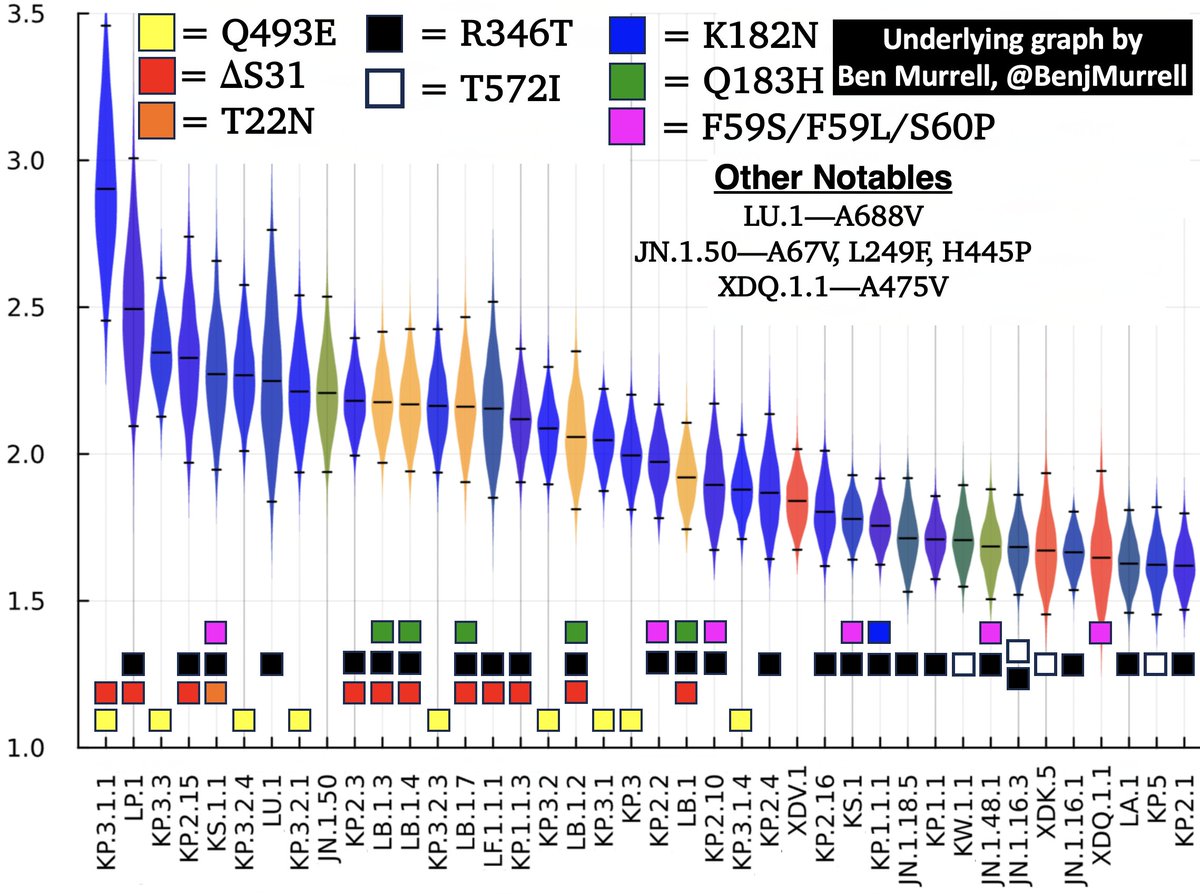
I’ve mostly pooh-poohed the rise of XEC for 2 reasons:
#1. Its spike is almost identical to the dominant KP.3.1.1
#2. I don’t think its advantage over KP.3.1.1 is large enough to make a significant real-world impact.
But one aspect of XEC is noteworthy: The demise of N*.
1/20
#1. Its spike is almost identical to the dominant KP.3.1.1
#2. I don’t think its advantage over KP.3.1.1 is large enough to make a significant real-world impact.
But one aspect of XEC is noteworthy: The demise of N*.
1/20
I’ve talked a lot about nucleocapsid before, mostly in this 120-tweet thread that was too long for anyone to want to read. Nucleocapsid is by far the most abundant SARS-CoV-2 protein. Nothing else comes close. It is very, very important. 2/20
https://x.com/LongDesertTrain/status/1781128579931128228
An essential aspect of N is its phosphorylation. Phosphorylation involves the attachment of a highly negatively charged phosphate to (usually) an S or T amino acid. We even know how it happens in the SARS-CoV-2 N. It’s pretty neat. 3/20
https://x.com/LongDesertTrain/status/1781140136723435712
N phosphorylation acts as a “molecular switch.” N is phosphorylated immediately after entry (possibly assisting the separation of the viral genome from N, with which it is intertwined and from which it must escape in order to be translated into proteins. 4/20 

The N3 region of the SARS-2 N is densely phosphorylated. N3 is highly conserved in betaCoVs, yet it’s been frequently mutated in SARS-2. Remarkably, the mutations in N3 invariably reduce phosphorylation—like the NSP3-N revertants I discussed here. 5/20
https://x.com/LongDesertTrain/status/1781140158844223877
The #1 role of N is to encapsidate the viral genome & tuck it inside the virion. N strongly binds RNA, & the viral genome wraps itself around N inside the virion. But phosphorylation dramatically reduces N’s RNA-binding ability. Phosphorylated N (pN) cannot encapsidate gRNA. 6/20 

An awesome recent study by @BlackledgeLab (lead author @MaiiaBotova) outlined one of the mechanisms underlying this phosphorylation “switch.” N3 phosphorylation causes it to bind to N2 in precisely the same way RNA does. 7/20





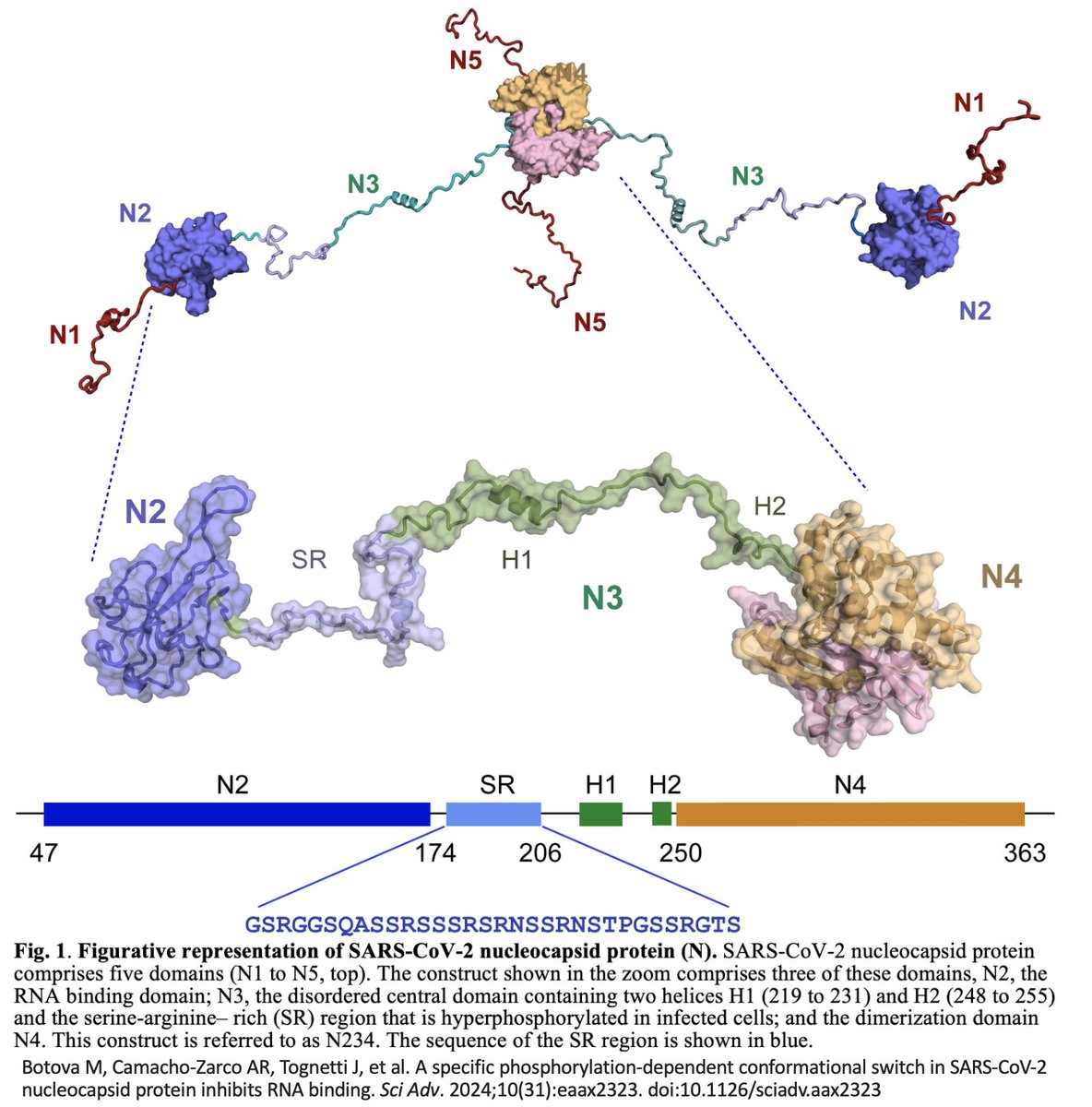
So why is N phosphorylated? Apart from a likely role in separating viral RNA from the genome upon cell entry, phosphorylation enables N to carry out its myriad other roles. Many proposed pN functions are uncertain or poorly understood, but a few are clearly important & real. 8/20
A fantastic preprint by a veritable all-star team—including @abdullah_m_syed, @doudna_lab, @KroganLab, @Doctor_Bou, & @TheOttLab—showed that phosphorylation improves N’s ability to facilitate RNA replication but impedes viral assembly. 9/20 

Similarly, the numerous mutations in the N3 region in SARS-2 variants, which reduce N3 phosphorylation, greatly improve viral assembly—by more than 10-fold. But reduced phosphorylation hampers RNA replication. So the virus faces a conflict. 10/20
Is there a way to have the best of both worlds? Indeed, the virus may have found a way: the novel N* protein. N* showed up very early in B.1.1. It involved a triple-nuc mutation that created a new TRS (see 🧵 below for TRS explanation). 11/20
https://x.com/LongDesertTrain/status/1673488117599285248
This new TRS results in the creation of a half-truncated version of N containing only its last half. Crucially, this half lacks the phosphorylated AA in the N3 region, meaning that it is not subjected to phosphorylation (& probably not harmful ISGylation either). 12/20
Incredibly, studies have found that despite lacking the first half of N—which contains the primary RNA-binding region—N* is capable of encapsidating the SARS-CoV-2 genome all by itself! 13/20




With N*, the virus could have its cake and eat it too. It can go ahead and phosphorylate N a bit more, boosting RNA synthesis, without having to sacrifice as much assembly efficiency since little N* can take up the slack on that front. 14/20
It’s little wonder, then, that multiple studies found that the N:R203K/G204R mutation that created N* resulted in higher viral loads and more severe illness. 15/20
https://x.com/LongDesertTrain/status/1745578167501263247
Finally, we come back to XEC. XEC is a recombinant of KP.3.3 and KS.1.1. When KP.3.3 first emerged, I thought it would not be around for long. Why? It has a mutation that basically destroys the N* TRS. It almost certainly produces ~zero N* protein. 16/20 

But to my surprise KP.3.3 has done very well, dominating in Japan, for example.
And XEC inherited this N*-destroying mutation from
KP.3.3.
17/20
And XEC inherited this N*-destroying mutation from
KP.3.3.
17/20

There’s no obvious reason XEC is doing as well as it is. Its spike is nearly identical to KP.3.1.1, & the few non-spike differences seem insignificant—except the N* destroyer. That XEC is doing well & lacks N* suggests N* is no longer advantageous. Could it be deleterious? 18/20
I really don’t understand why this should be. The only change in N in JN.1-descended lineages is Q229K. This is near the end of the crucial leucine-rich helix. Could it somehow have made N* superfluous? Seems unlikely but who knows? 19/20 

Something has changed, I know not what. But I think it’s worth taking note of. To me, this is the most interesting part of XEC’s recent success.
The meaning of the (possible) demise of N* isn't clear, but perhaps it holds a clue to some deeper mechanism at work.
20/20
The meaning of the (possible) demise of N* isn't clear, but perhaps it holds a clue to some deeper mechanism at work.
20/20
• • •
Missing some Tweet in this thread? You can try to
force a refresh