With the XEC variant on the way to dominance in most places, and XEC waves starting to show in COVID metrics, it is time to ponder which variant will drive the next wave after XEC.
Here are the leading contenders: LP.8.1, MC.10.2, MV.1, and XEC.4.
🧵
Here are the leading contenders: LP.8.1, MC.10.2, MV.1, and XEC.4.
🧵

They are shown above using a log scale, so you can compare their growth rates vs OG XEC.
🧵
🧵
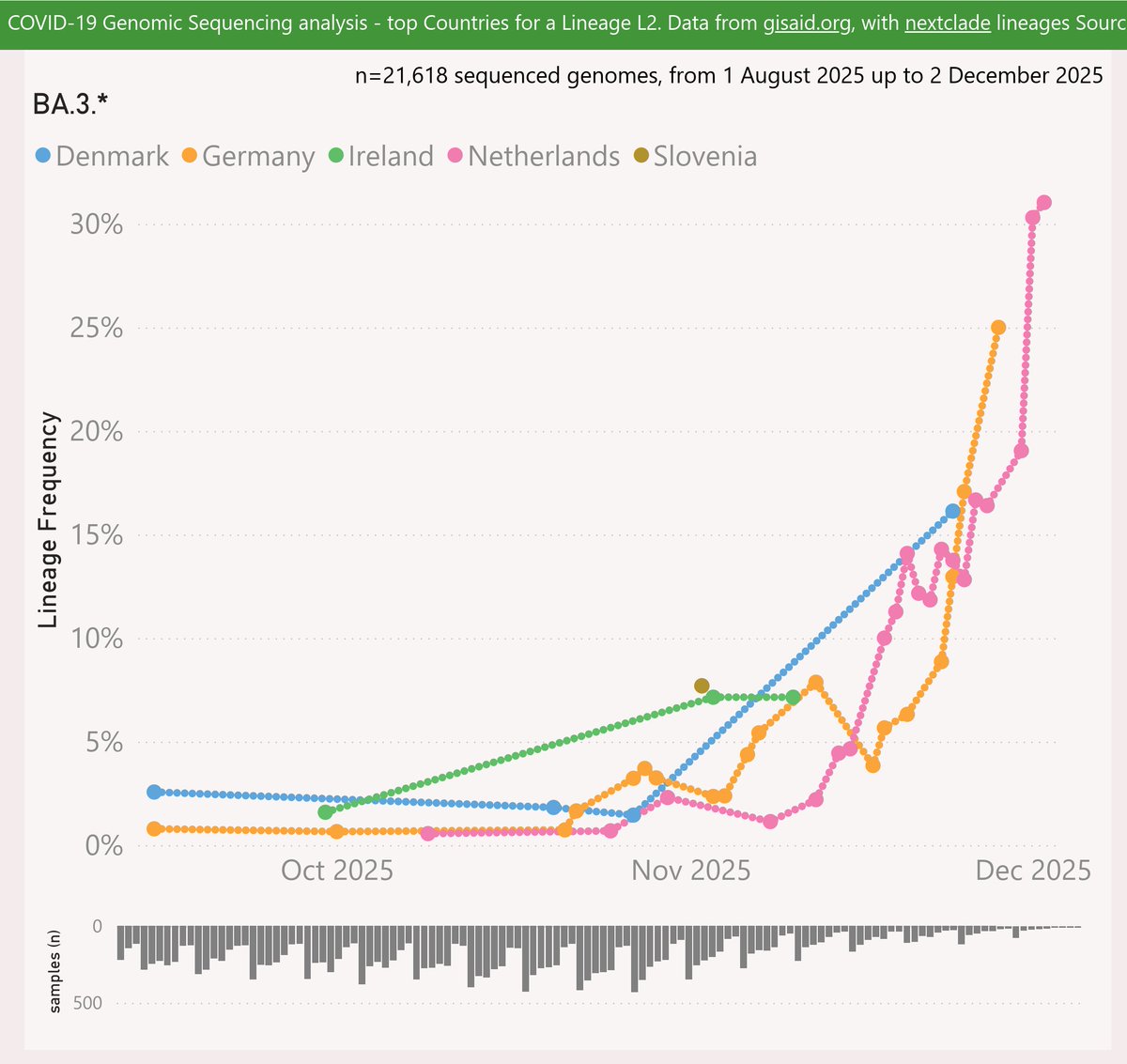
LP.8.1 is descended from FLiRT KP.1.1.3. LP.8 adds the Spike:Q493E mutation (featured in FLuQE) and also H445R and F186L – a potent example of convergent evolution. Then LP.8.1 adds R190S.
LP.8.1 has had some success in Taiwan, Japan, Singapore and lately in the US.
🧵
LP.8.1 has had some success in Taiwan, Japan, Singapore and lately in the US.
🧵
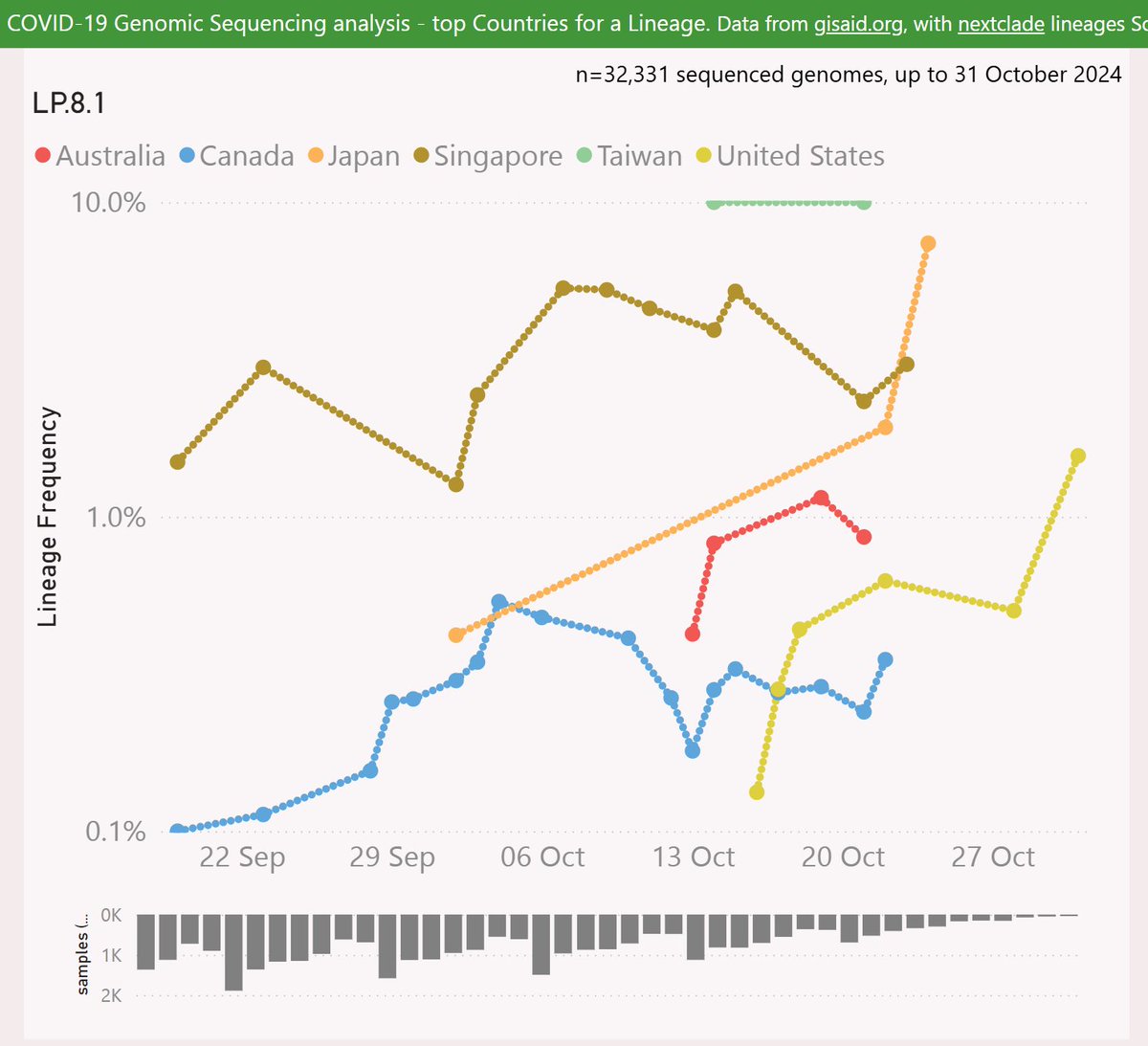
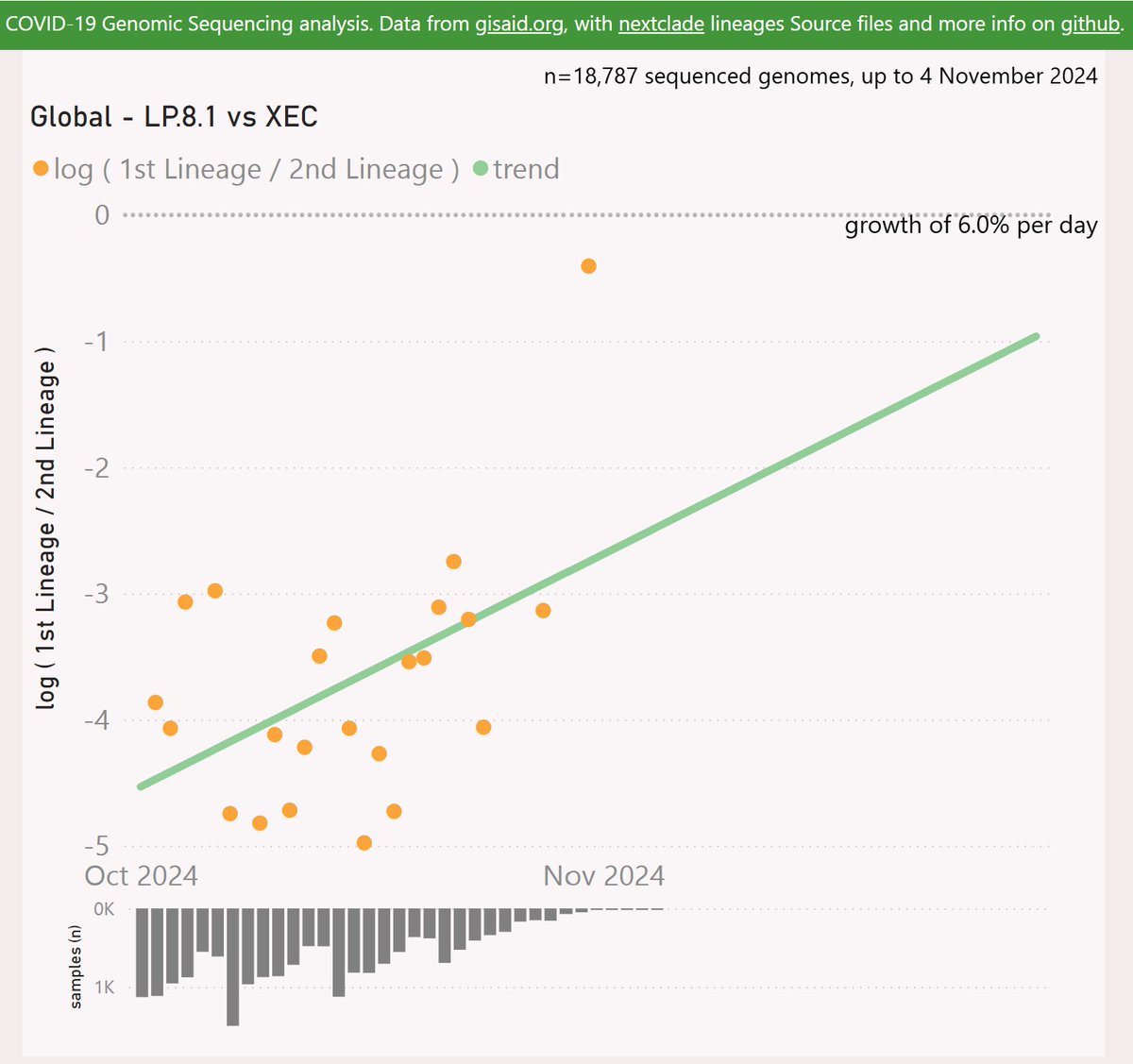
Globally, LP.8.1 is showing a growth advantage of 6% per day (42% per week) vs XEC, so any crossover looks like happening in December.
🧵
🧵
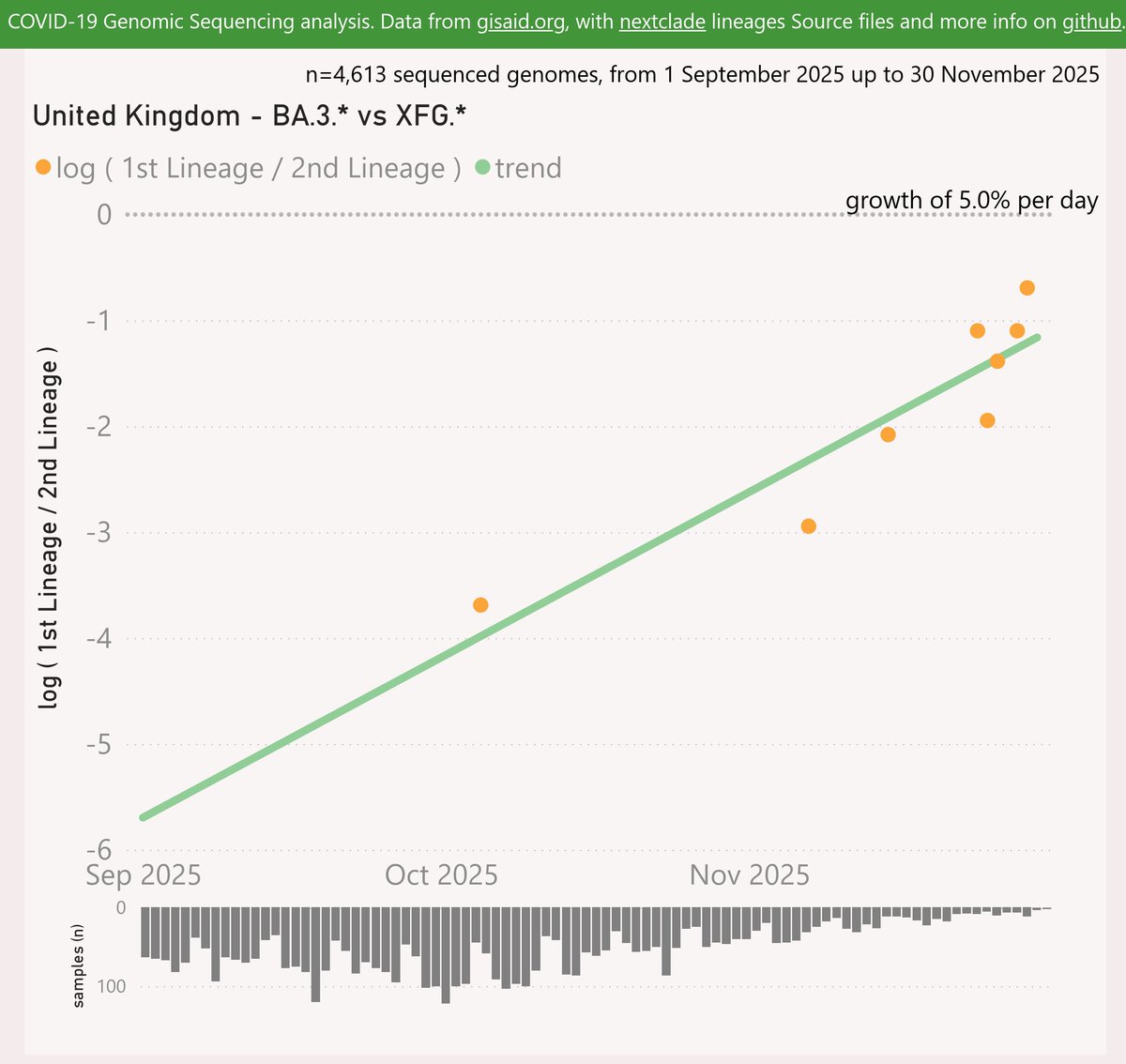
MC.10.2 is descended from DeFLuQE KP.3.1.1, so boosts the resistance of the dominant DeFLuQE clan. MC.10.2 adds the Spike I101T and A376D mutations.
MC.10.2 has had some success in NZ and Ontario, Canada.
🧵
MC.10.2 has had some success in NZ and Ontario, Canada.
🧵
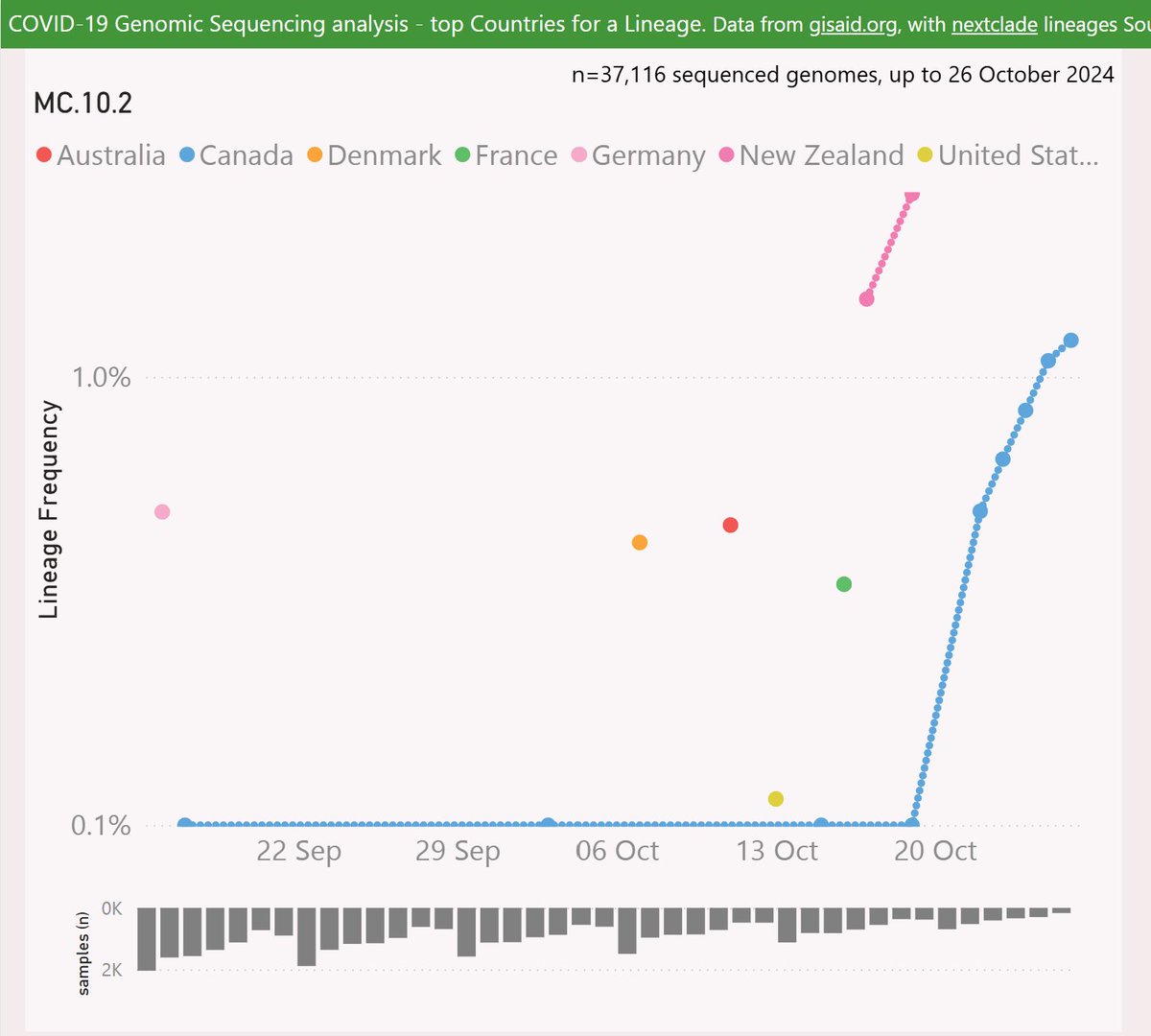
Before you say “oh Ontario huh, just like XEM”, please note that there was a wide geographic distribution before it took hold there.
🧵
🧵
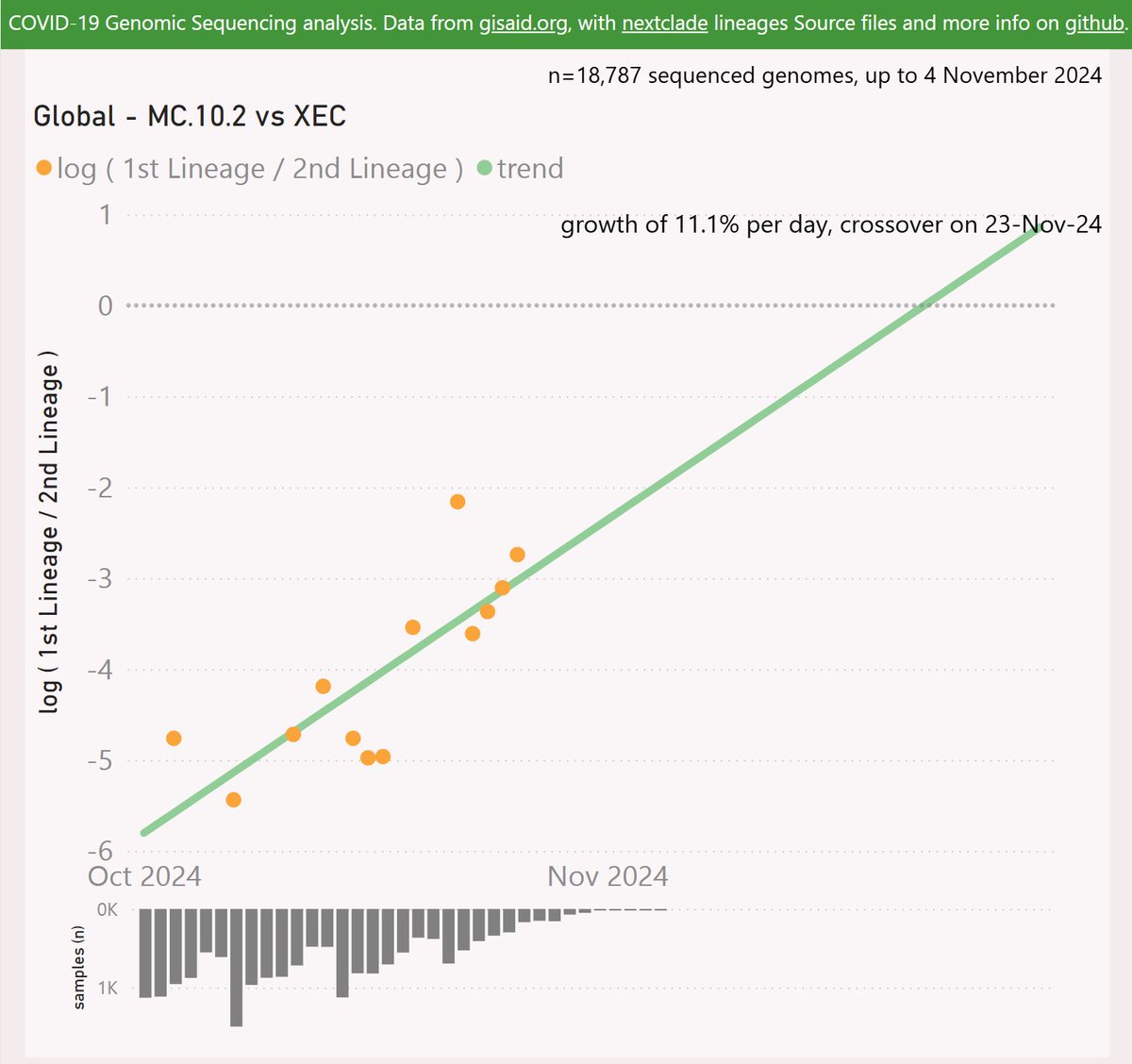
Globally, MC.10.2 is showing a striking growth advantage of 11.1% per day (78% per week) vs XEC, which predicts a crossover in late November.
🧵
🧵
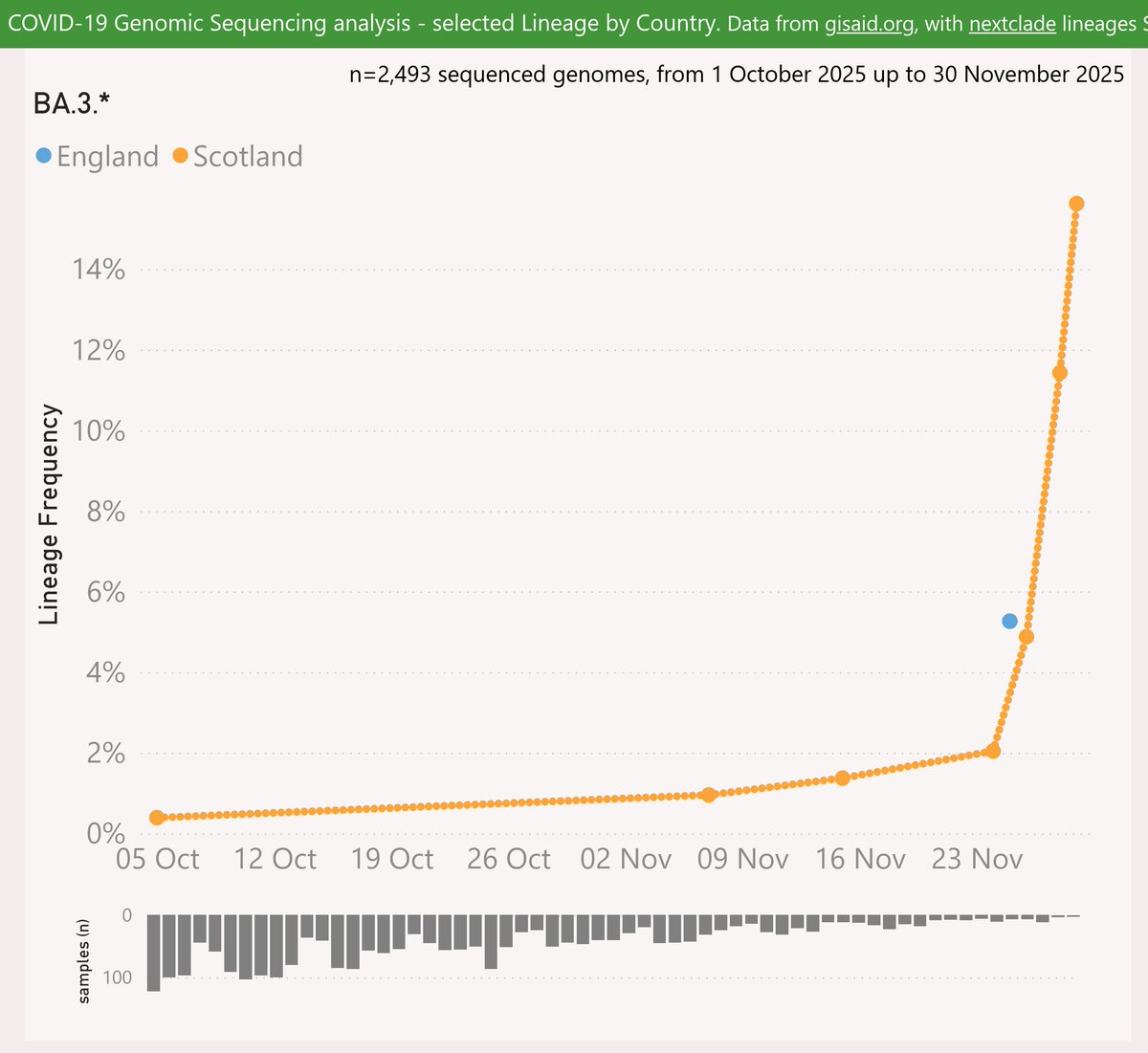
MV.1 is descended from JN.1.49.1 via MB.1.1.1. MV.1 adds the Spike:K478T mutation.
Singapore is at around 50% frequency, and it seems quite prevalent in Mauritius. It is also growing strongly in Australia and the US.
🧵
Singapore is at around 50% frequency, and it seems quite prevalent in Mauritius. It is also growing strongly in Australia and the US.
🧵
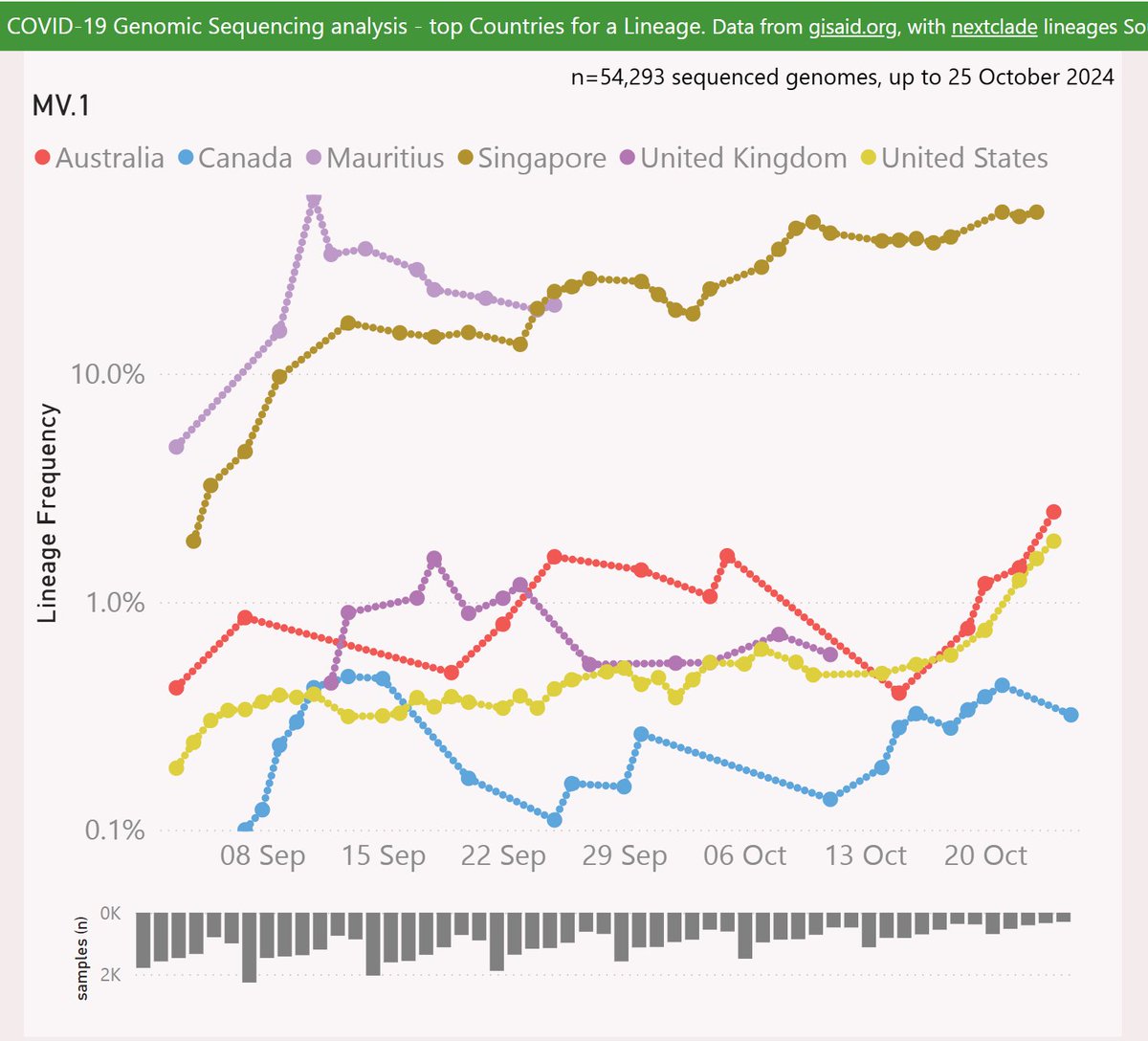
Globally, MV.1 is showing an accelerating growth advantage of 3.5% per day (25% per week) vs XEC, so any crossover looks like December or later.
🧵
🧵

XEC.4 adds the Spike T572I mutation.
XEC.4 has been most successful in New Zealand and Australia, and is also growing strongly in Sweden, the UK and Canada.
🧵
XEC.4 has been most successful in New Zealand and Australia, and is also growing strongly in Sweden, the UK and Canada.
🧵

Globally, XEC.4 is showing an accelerating growth advantage of 4.9% per day (34% per week) vs XEC, so any crossover looks like December or later.
🧵
🧵
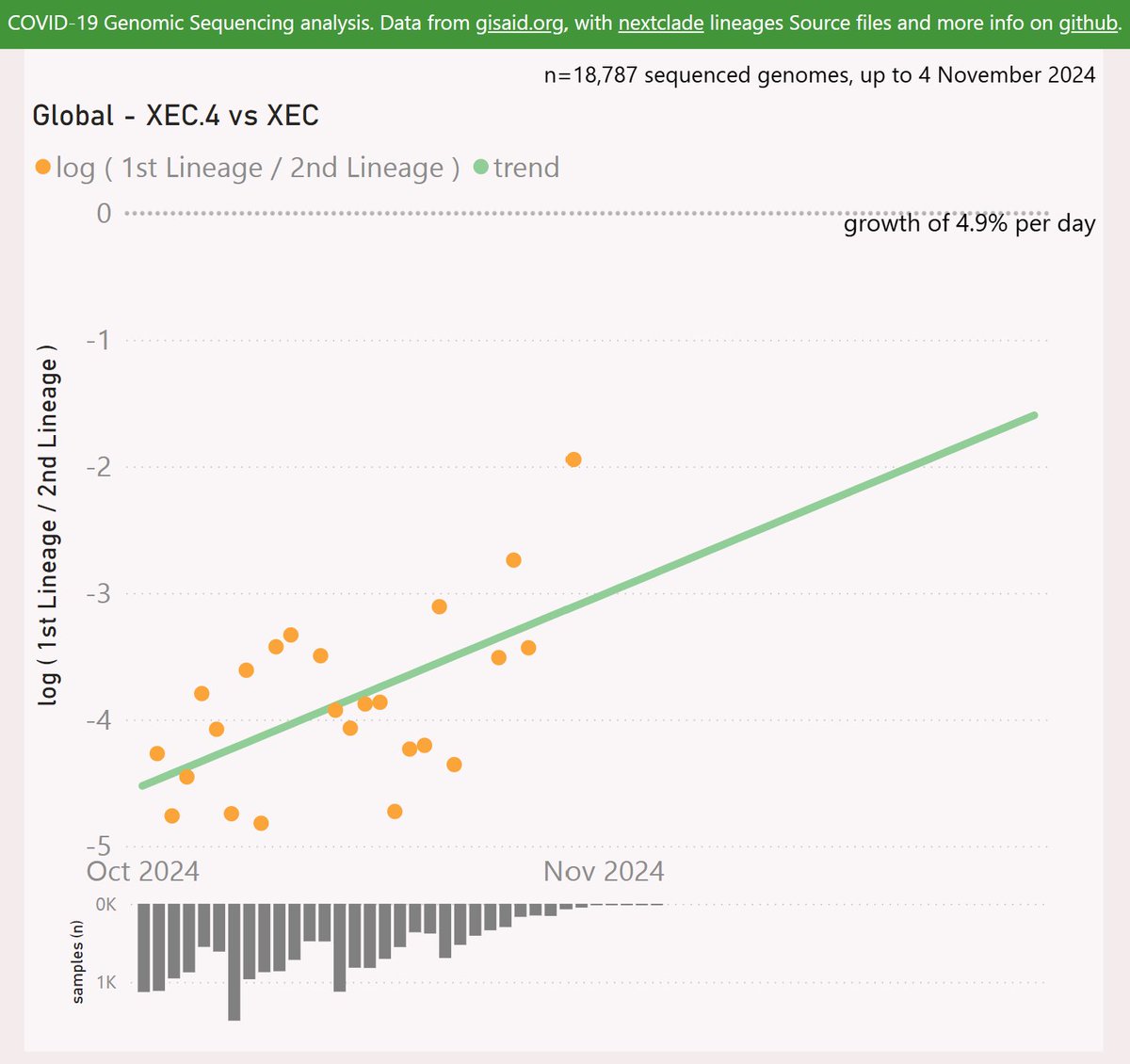
So in summary, the most obvious contenders to challenge XEC at this point are MV.1, LP.8.1 and MC.10.2. I will continue to back MV.1, with it's high growth rate and it's success in undersampled regions.
I will continue to monitor the other contenders.
🧵
I will continue to monitor the other contenders.
🧵

The usual caveats apply - recent sample sizes are smaller which might skew these results, and “global” sequencing data is dominated by wealthy countries, with many under-sampled regions.
🧵
🧵
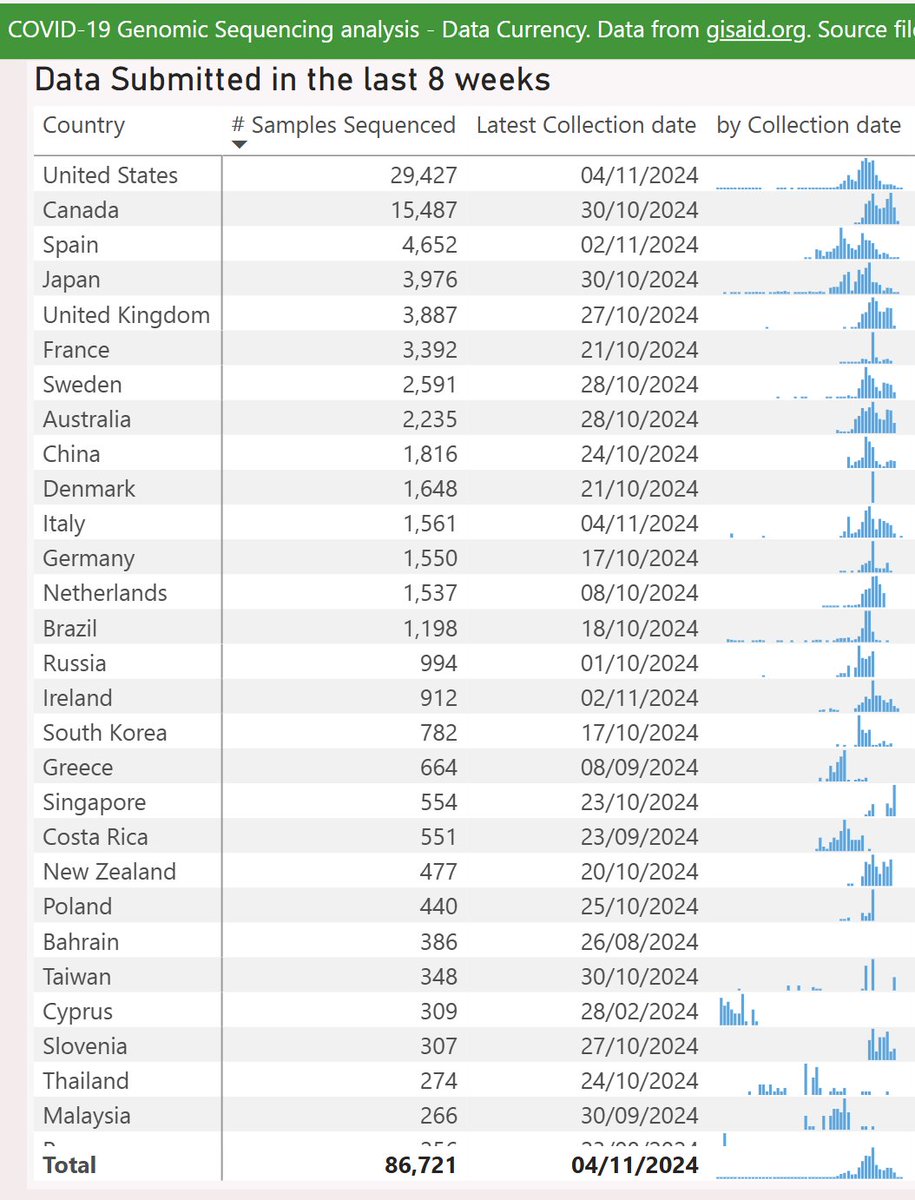
Interactive genomic sequencing dataviz, code, acknowledgements and more info here:
🧵 endsgithub.com/Mike-Honey/cov…
🧵 endsgithub.com/Mike-Honey/cov…
• • •
Missing some Tweet in this thread? You can try to
force a refresh