I am very skeptical of the AI/ML/computational methods to predict protein-protein interactions. Prove me wrong.
I present a challenge for anyone who claims they can predict protein-protein interactions.
1/n
I present a challenge for anyone who claims they can predict protein-protein interactions.
1/n
We are gearing up to post a big paper on de novo protein binder discovery. As part of that we generated lots of data on both novel binders (mini protein scaffolds) and non-binders. For example... 2/n
Here is binding data: 15 novel binders each bind one of 15 target proteins selectively (with LC3B/GABARAP crosstalk, expected due to similarity). This is real data. Now, lets see what @googledeepmind alphafold3 predicts should bind within this 15x15 potential array of PPIs... 3/n 
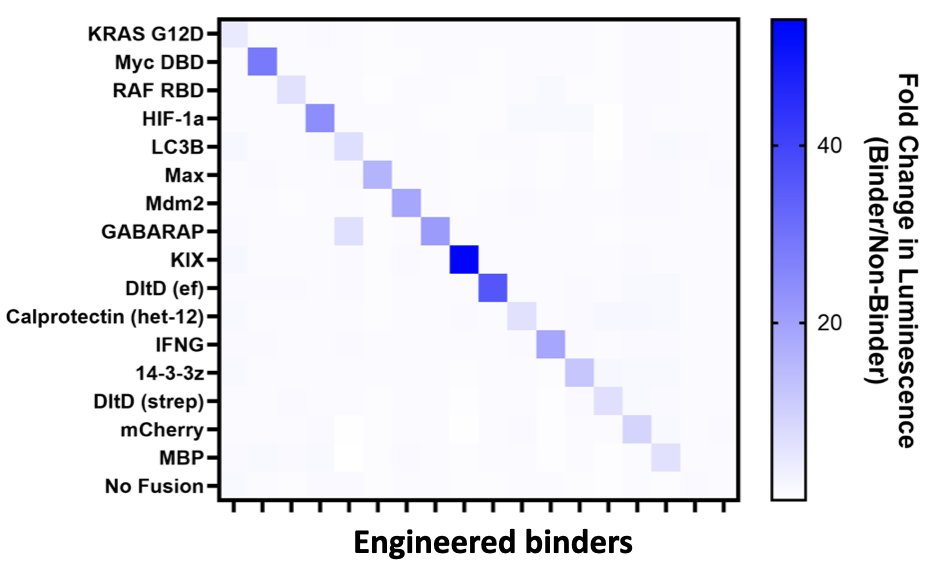
Here is the Alphafold3 prediction. It is worse than random. High false positive and high false negative (iPTM/PTM shown - other metrics look very similar). 4/n 

So, if you or someone you know thinks they can actually predict PPIs, prove it. Here is a link to our blinded protein sequence data:
5/ndickinsonlab.uchicago.edu/ppi-challenge
5/ndickinsonlab.uchicago.edu/ppi-challenge
Your task - Match up the real PPIs. Email me the answer at dickinson@uchicago.edu and I'll tell you if you are correct. If so, we have thousands (millions) more test cases to prove out your approach! 6/n
There is clearly lots of work to do in AI/ML-based interaction detection. Hopefully our datasets help in the future - but there are so many high profile PPI detections papers popping up recently. I'm a skeptic. Someone prove me wrong! 7/n
• • •
Missing some Tweet in this thread? You can try to
force a refresh




