BREAKING🔔 A new study from G2P-Japan🇯🇵, led by Keiya @Keiya717, is out at @biorxivpreprint. We assessed the humoral immunity induced by JN.1 mRNA vaccines against a broad range of SARS-CoV-2 variants including JN.1, KP.3.1.1 & XEC. Please repost🔥 1/
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
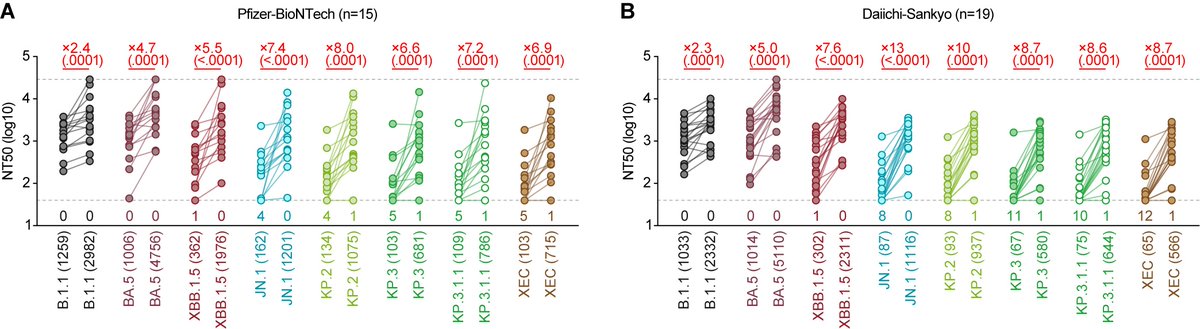
In this study, we used 2 mRNA vaccines, one is from Pfizer/BioNTech🇺🇸🇩🇪, while another is from Daiichi-Sankyo🇯🇵. The good news is that there are no significant differences between them and both broadly neutralized a variety of SARS-CoV-2 including JN.1, KP.3.1.1 and XEC. 2/ 

But there are two scientific interests.
First, compared to our recent study using JN.1 and KP.3.3 breakthrough (i.e. natural) infection sera, antiviral humoral immunity against KP.3 and XEC was weakly induced. However, JN.1 mNRA vaccine can. Why🤔?? 3/
First, compared to our recent study using JN.1 and KP.3.3 breakthrough (i.e. natural) infection sera, antiviral humoral immunity against KP.3 and XEC was weakly induced. However, JN.1 mNRA vaccine can. Why🤔?? 3/

FYI - we have reported the findings on the efficiency of antiviral humoral immunity induced by the natural infection of JN.1 and KP.3.3 in Lancet Infectious Diseases @TheLancetInfDis. 4/
https://x.com/G2PJapan/status/1855045437612515731
Second, we used the XBB.1.5 mRNA vaccine sera last year. Its antiviral humoral immunity against ancestral B.1.1 was greater than that against XBB.1.5. 5/ 

However, the JN.1 mRNA vaccine induced a more pronounced antiviral effect against XBB.1.5 than against ancestral B.1.1. Again, why🤔??? 6/ 

*In figures, the numbers in parentheses indicate the 50% neutralization titer of each serum. Please compare these values between the figures.
Our previous report on XBB.1.5 monovalent mRNA vaccine is also in Lancet Infectious Diseases @TheLancetInfDis. 7/
Our previous report on XBB.1.5 monovalent mRNA vaccine is also in Lancet Infectious Diseases @TheLancetInfDis. 7/
https://x.com/SystemsVirology/status/1744582373289914483
These observations imply that immune imprinting has shifted from that biased towards pre-Omicron to that biased towards Omicron, depending on the time and/or number of immune stimuli (e.g., infection and/or vaccination)🤔???
Good news, and thought provoking🤔 8/8
Good news, and thought provoking🤔 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh