This is worth a full read.
They really knocked it out of the park on this study.
publichealthpolicyjournal.com/biontech-rna-b…
They really knocked it out of the park on this study.
publichealthpolicyjournal.com/biontech-rna-b…
First…
They measure how long is spike expressed in cells.
7 days out it is higher than day 1. Peaks at day 5.
They measure how long is spike expressed in cells.
7 days out it is higher than day 1. Peaks at day 5.
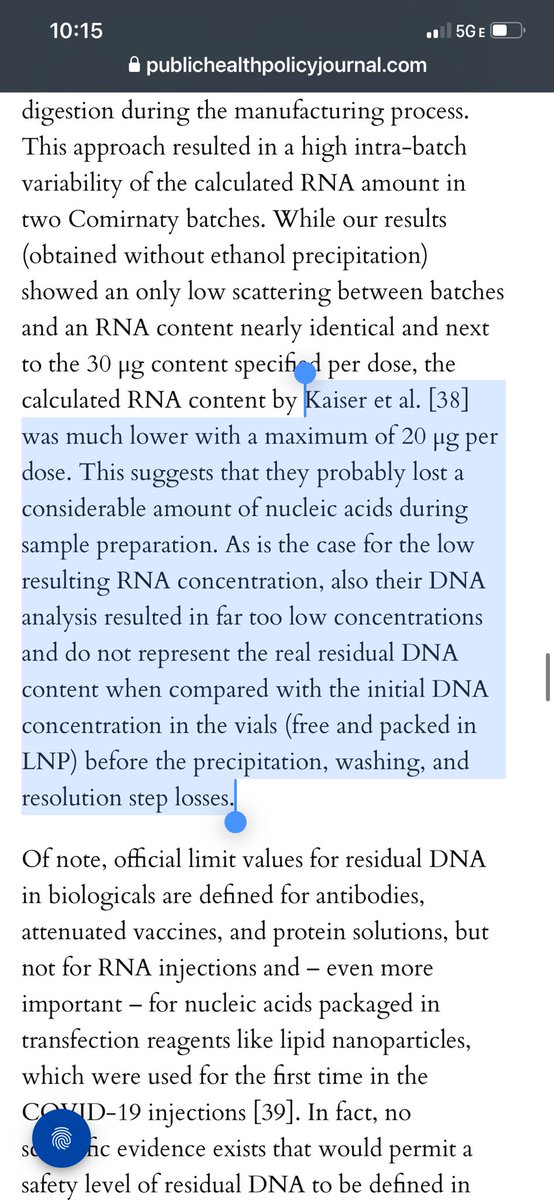
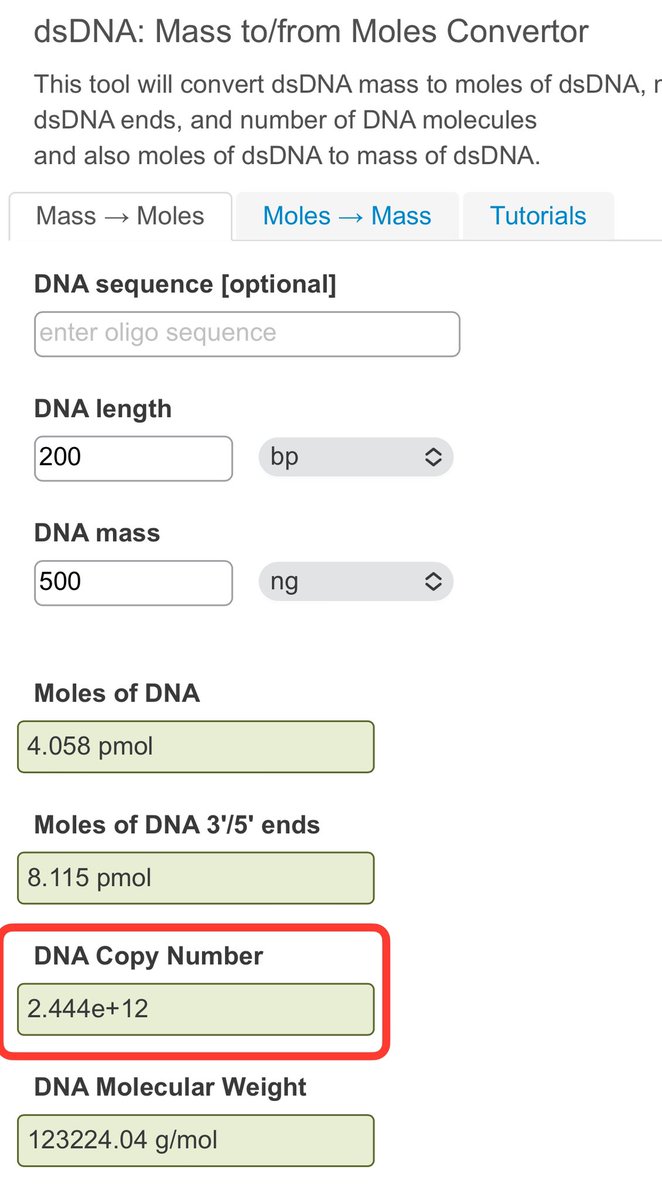
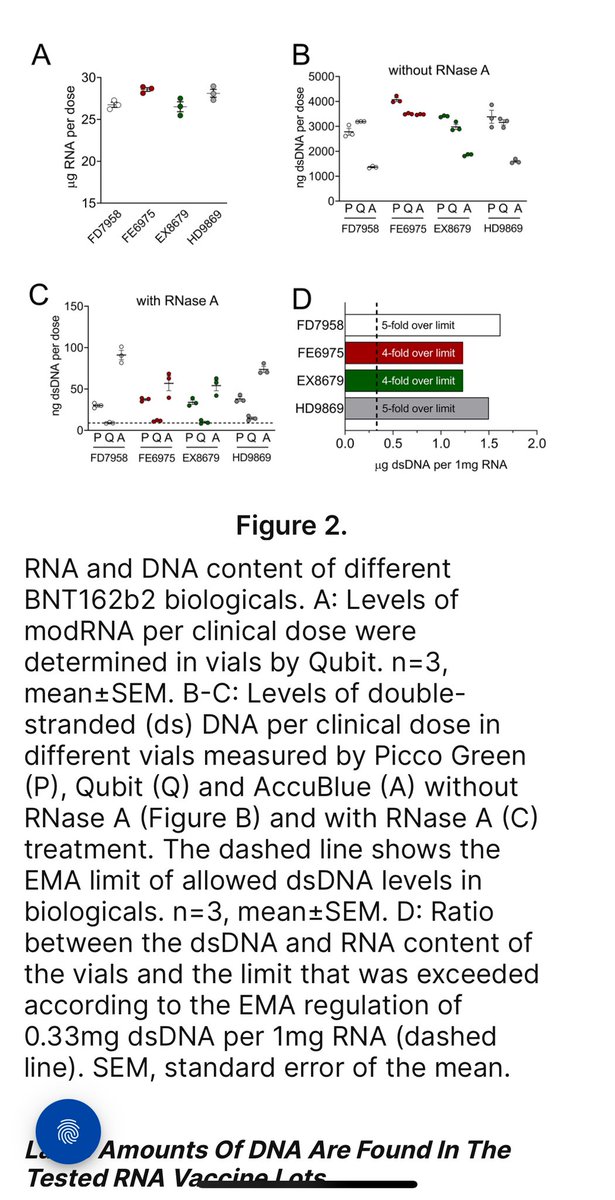
Using 3 different Fluorometry methods they zero in on DNA levels before and after RNaseA treatments.
4-5X over once RNA is removed.
The AccuBlue dyes had the least cross talk.
4-5X over once RNA is removed.
The AccuBlue dyes had the least cross talk.
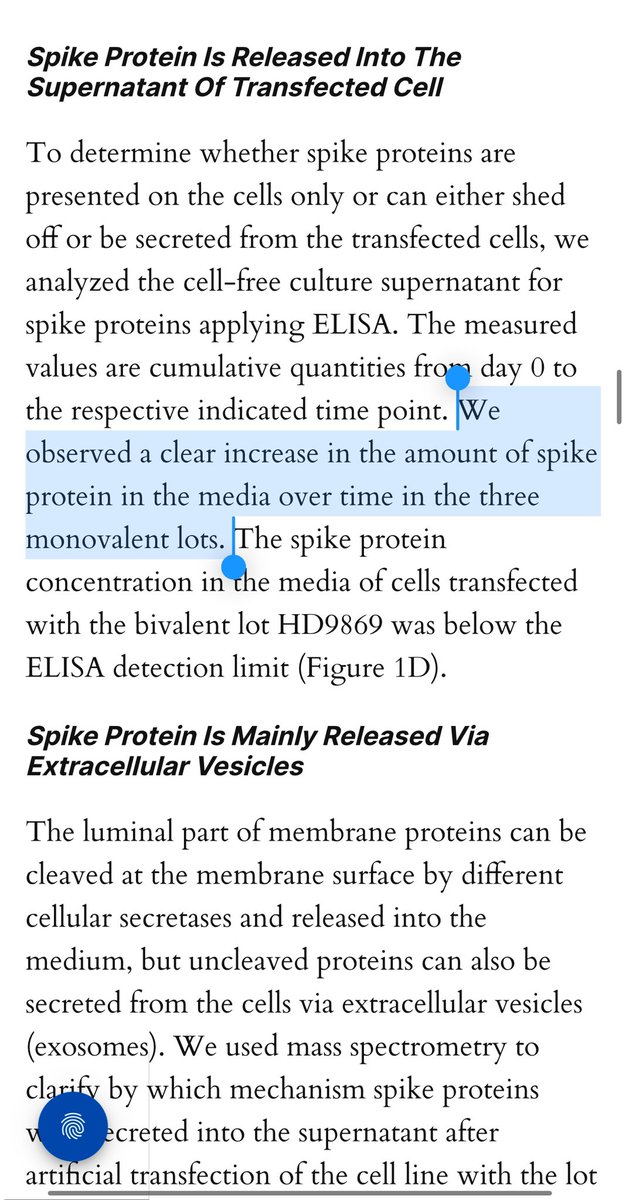
Most importantly they show the spike is packaged in exosomes which likely renders most ELISA assays used to track this somewhat blind to quantitation and begs the question of shedding since exosomes are exhaled and secreted in the skin.
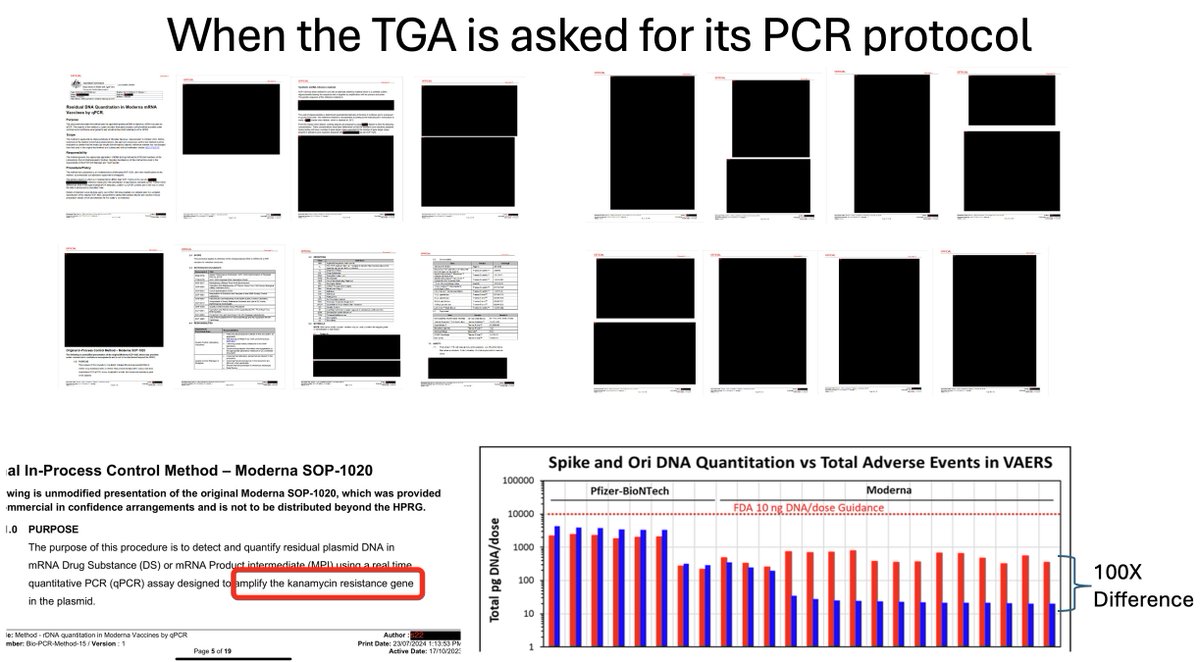
@TGAgovau are you awake yet?
@TGAgovau are you awake yet?
A very good discussion on SV40 and its reckless use given its known functionality.
Forward this paper to all politicians and regulators.
@FLSurgeonGen @DrJBhattacharya @MartyMakary @SenatorRennick @RobertKennedyJr @RWMaloneMD @BroadbentMP @P_McCulloughMD @MendenhallFirm @Double_Christ
Forward this paper to all politicians and regulators.
@FLSurgeonGen @DrJBhattacharya @MartyMakary @SenatorRennick @RobertKennedyJr @RWMaloneMD @BroadbentMP @P_McCulloughMD @MendenhallFirm @Double_Christ
The review actually started in July so sailing through is probably a misnomer.
And the cat photo is from nightcafe AI.
They still seem to have problems with hands and paws!
They still seem to have problems with hands and paws!
Just to connect the dots on the exosomes.
This would explain shedding and if any of these exosomes have replication competent plasmids, we have a mess on our hands.
The Beck et al work likely also has fecal/oral routes of transmission.
The Karen Konclave may start buying masks again:)
This would explain shedding and if any of these exosomes have replication competent plasmids, we have a mess on our hands.
The Beck et al work likely also has fecal/oral routes of transmission.
The Karen Konclave may start buying masks again:)

• • •
Missing some Tweet in this thread? You can try to
force a refresh