I’m pleased to share that we FINALLY submitted our latest manuscript on SARS-CoV-2 cryptic lineages and what they tell us about the origins of COVID-19.
This was a ton of work.
1/medrxiv.org/cgi/content/sh…
This was a ton of work.
1/medrxiv.org/cgi/content/sh…
First, standard background. Cryptic lineages are unique, evolutionarily advanced SARS-CoV-2 lineages detected from wastewater.
We are fairly certain that these lineages come from individuals with very long infections (not animals).
2/
We are fairly certain that these lineages come from individuals with very long infections (not animals).
2/
https://x.com/SolidEvidence/status/1587551682388295680
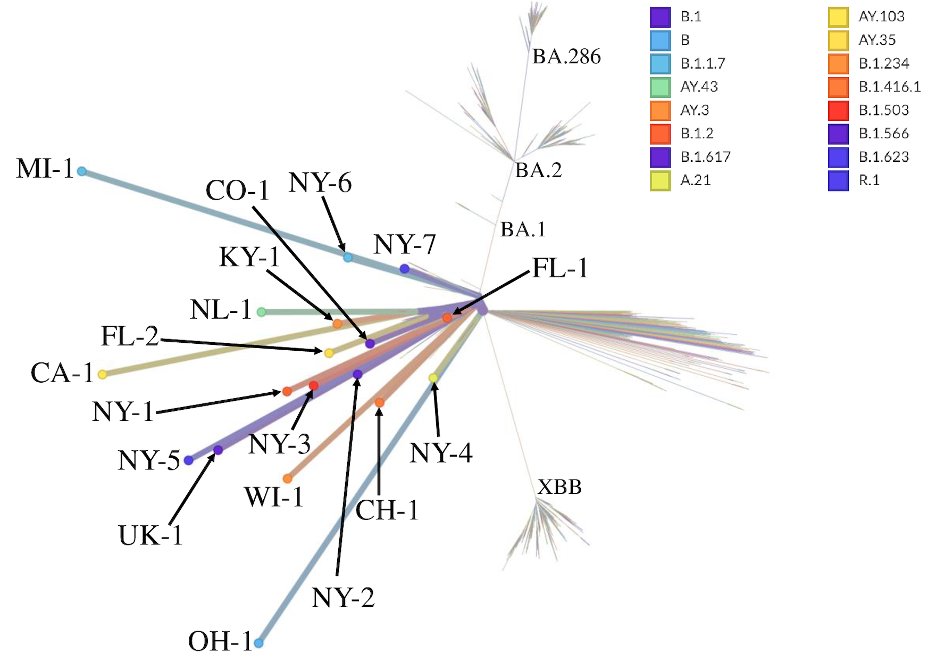
We’ve had several previous papers on cryptic lineages, but this new manuscript is about lineages we found in public sequence databases.
We screened the raw data from 135,672 wastewater samples from over 2k sites across 45 countries collected prior to November, 2023.
3/
We screened the raw data from 135,672 wastewater samples from over 2k sites across 45 countries collected prior to November, 2023.
3/
The first part of the manuscript is just about the methods we used for finding and characterizing these lineages.
I won’t get into the details, but it’s straightforward. We look for things that appear reproducibly but do not match known lineages.
4/
I won’t get into the details, but it’s straightforward. We look for things that appear reproducibly but do not match known lineages.
4/

In total we found 18 cryptic lineages. 6 had been described before (3 by us, 3 by others), the rest were novel.
The lineages were all anachronistic (out of time), meaning they were all derived from lineages that had stopped circulating long before the cryptic was detected.
5/
The lineages were all anachronistic (out of time), meaning they were all derived from lineages that had stopped circulating long before the cryptic was detected.
5/
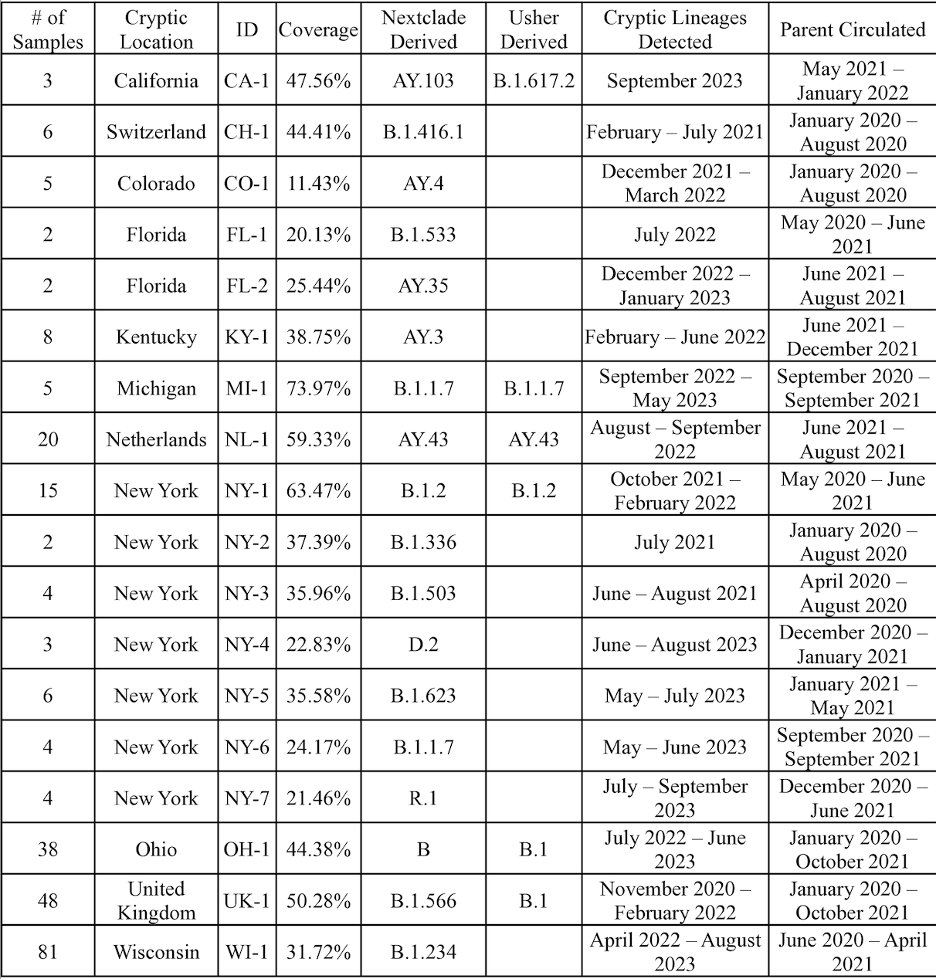
One lineage worth particular note was the Ohio (OH-1) lineage, which we detected regularly from two different sewersheds that were 40 miles apart.
7/
7/
In addition to following the sequences submitted to SRA, was also got wastewater samples from both sewersheds and tracked the lineage independently.
8/
8/
The lineage persisted until summer 2023 when it spiked and then abruptly disappeared.
I doubt it ended well, but we will probably never know.
9/
I doubt it ended well, but we will probably never know.
9/
As an aside, the Ohio lineage was a big deal for us. It was the first time we identified a cryptic lineage from a public database, tracked down the sewershed, got them to send us wastewater, and found the exact same lineage.
This was a good sanity check.
10/
This was a good sanity check.
10/
An interesting point about the OH-1 lineage is it picked up new mutations over time, and when a new mutation appeared, it appeared in BOTH sewersheds.
The best explanation for this is that the lineage came from a single source that contributed to both sewersheds.
11/
The best explanation for this is that the lineage came from a single source that contributed to both sewersheds.
11/
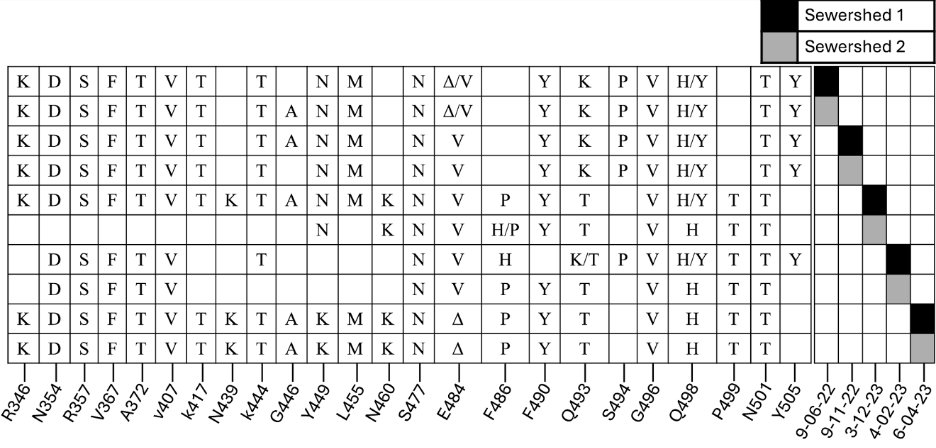
Another interesting point. We identified 5 different insertions in the cryptic lineages. 4 were derived from SARS-CoV-2 sequence, 1 was not.
This is just a reminder that intra- and inter- sequence recombination leading to insertions is a common practice in Coronaviruses.
12/
This is just a reminder that intra- and inter- sequence recombination leading to insertions is a common practice in Coronaviruses.
12/
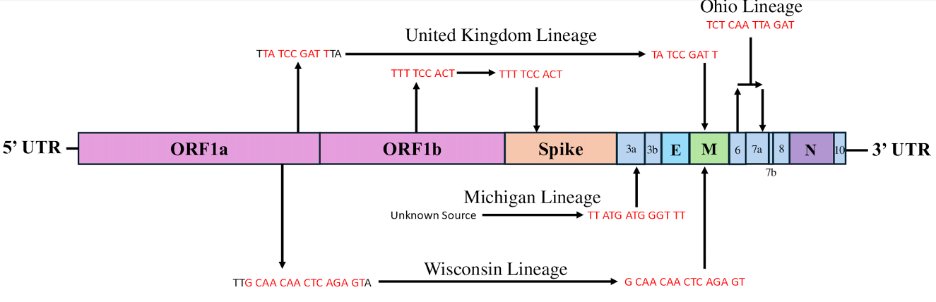
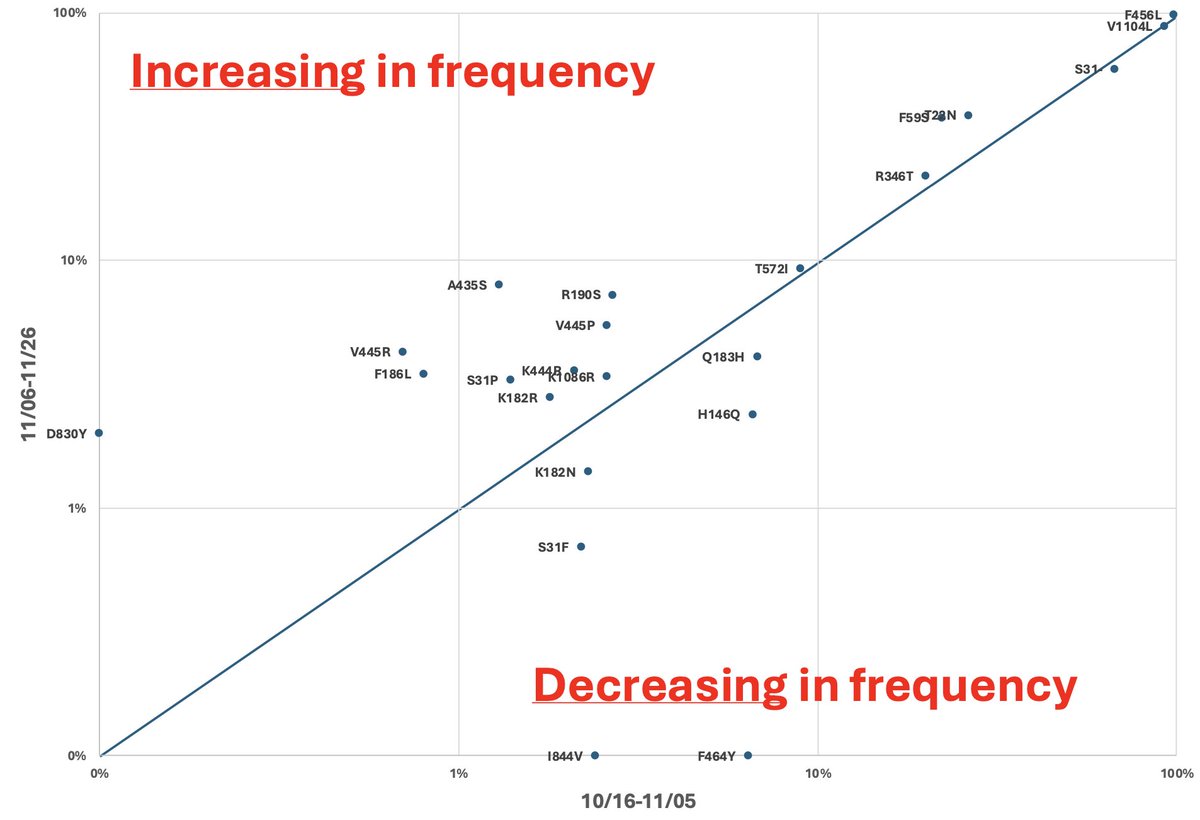
There was an impressive degree of convergent evolution among the 18 cryptic lineages.
There were 83 mutations that were observed in at least 3 of the lineages and 79 of these changed a protein sequence.
13/
There were 83 mutations that were observed in at least 3 of the lineages and 79 of these changed a protein sequence.
13/
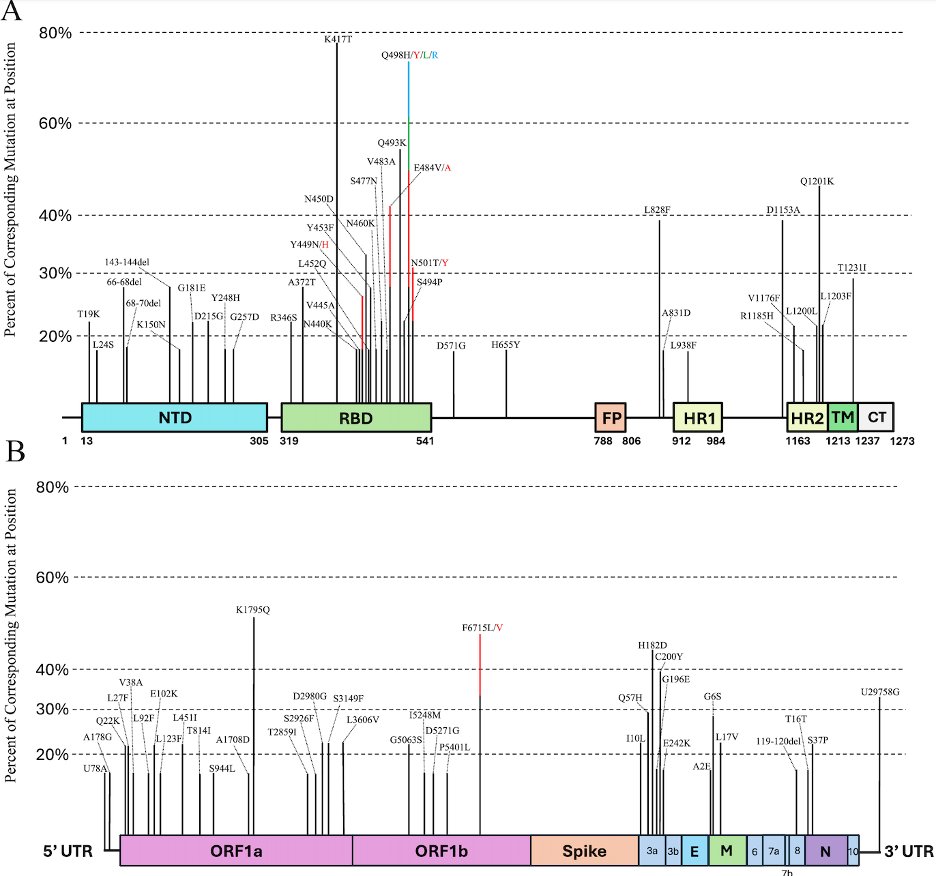
One thing we noticed is that many of the most common convergent mutations changed the sequence to match that of SARS-CoV-2’s closest relative from bats, RaTG-13.
14/
14/
To explore this further, we tabulated all of the amino acid positions that were conserved among closely related Sarbecoviruses but changed in SARS-CoV-2.
There were a total 26 SARS-CoV-2-specific changes.
15/
There were a total 26 SARS-CoV-2-specific changes.
15/
Amazingly, 12/26 of these positions had reverted to the Sarbeco consensus sequence in at least 1 cryptic lineage, and 7 of them had reverted in at least 3 cryptic lineages.
Two of the reversions appeared in HALF of the cryptic lineages. This is astounding.
16/
Two of the reversions appeared in HALF of the cryptic lineages. This is astounding.
16/
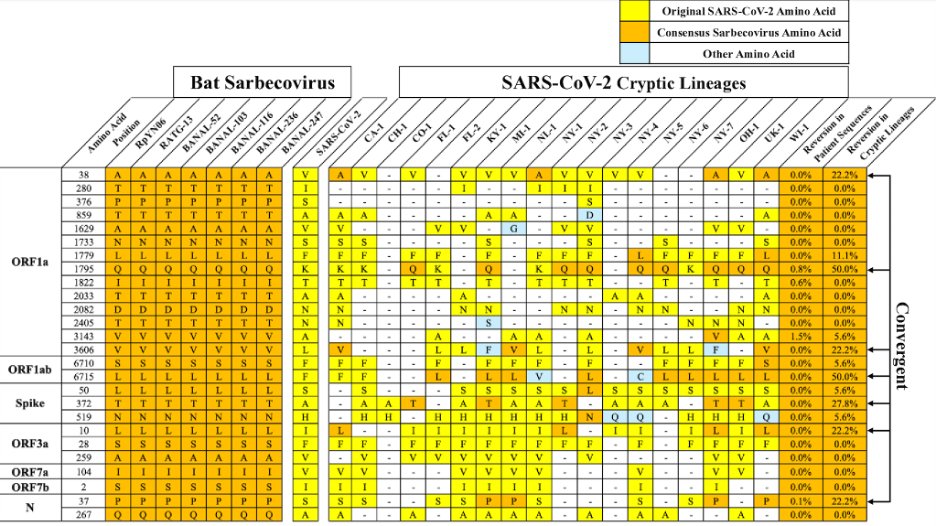
Why is this important?
This suggests that there is strong selective pressure on cryptic lineages to revert to the consensus Sarbecovirus sequence.
This is one reason we think cryptic lineages replicate in an environment similar to their bat ancestors (the GI tract).
17/
This suggests that there is strong selective pressure on cryptic lineages to revert to the consensus Sarbecovirus sequence.
This is one reason we think cryptic lineages replicate in an environment similar to their bat ancestors (the GI tract).
17/
What does this say about SARS-CoV-2 origins? It suggests that SARS-CoV-2 was replicating in a non-GI (likely respiratory) environment for a considerable period of time (probably years) prior to the start of the COVID pandemic.
18/
18/
This data presented DOES NOT say whether SARS-CoV-2 was zoonotic or from a lab. However, it does add some nuance to either scenario.
19/
19/
If SARS-CoV-2 was indeed a zoonosis, it probably did not come directly from a bat. It is much more likely that it had been circulating in the respiratory tract of an animal, such as one of the animals that we now know were present at the seafood market.
20/
20/
Alternatively, if SARS-CoV-2 came from a lab, it almost certainly was passaged for a long time prior to leaking and was not simply engineered ‘from scratch’.
21/
21/
The combination of mutations acquired by SARS-CoV-2 would not have been tolerated in its normal environment, but they are also not mutations that one would deliberately engineer.
22/
22/
One could argue that the virus was engineered, such as by adding a furin cleave site (FCS), and then passaged. However, the FCS could just as well have been introduced by a random insertion, as this manuscript demonstrates.
23/
23/
If the FCS were added and then passaged, it is hard to explain why the virus did not acquire D614G or enhance the FCS, which both quickly occurred upon spread in humans.
24/
24/
Either way, this data demonstrates that SARS-CoV-2 wasn’t a typical bat Sarbecovirus that jumped straight to humans; the mutations would not have been tolerated in that environment.
SARS-CoV-2 had been practicing for a while.
25/25
SARS-CoV-2 had been practicing for a while.
25/25
• • •
Missing some Tweet in this thread? You can try to
force a refresh