BREAKING🔔 The 50th🎉 paper from G2P-Japan🇯🇵 is out at Lancet Infectious Diseases
@TheLancetInfDis. We elucidated the virological characteristics of SARS-CoV-2 LP.8.1. Please repost🔥 1/
thelancet.com/journals/lanin…
@TheLancetInfDis. We elucidated the virological characteristics of SARS-CoV-2 LP.8.1. Please repost🔥 1/
thelancet.com/journals/lanin…
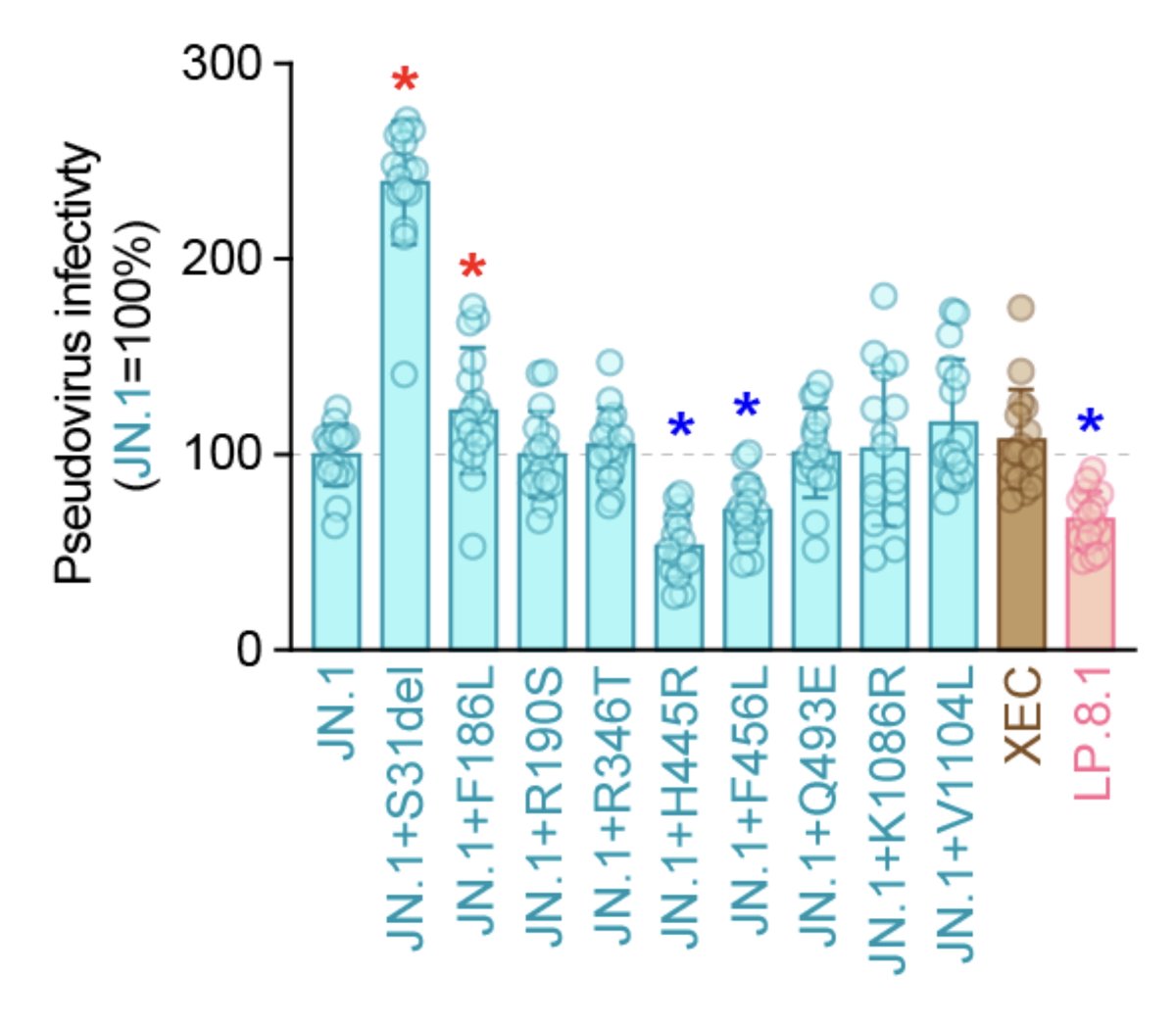
In spike, LP.8.1 has 9 mutations (S31del, F186L, R190S, R346T, V445R, F456L, Q493E, K1086R, and V1104L) compared with JN.1; and 8 amino acid residues (T22N, S31del, F59S, F186L, R190S, R346T, H445R, and K1086R) differ between the spike proteins of LP.8.1 and XEC. 2/ 

Transmissibility/fitness (Re):
Epidemic dynamics modeling led by Jumpei @jampei2 and Kaho showed that the Re of LP.8.1 is ~1.1-fold higher than that of XEC in 🇺🇸. However, the Re of LP.8.1 and XEC were almost comparable in 🇯🇵. Difference between countries🤔?? 3/
Epidemic dynamics modeling led by Jumpei @jampei2 and Kaho showed that the Re of LP.8.1 is ~1.1-fold higher than that of XEC in 🇺🇸. However, the Re of LP.8.1 and XEC were almost comparable in 🇯🇵. Difference between countries🤔?? 3/

Pseudovirus infectivity:
LP.8.1 < XEC = JN.1. Difference between our data (figure) and that from Yunlong @yunlong_cao (url link below) might be due to difference in experimental setup (pseudovirus vs RBD)🤔?? 4/
biorxiv.org/content/10.110…
LP.8.1 < XEC = JN.1. Difference between our data (figure) and that from Yunlong @yunlong_cao (url link below) might be due to difference in experimental setup (pseudovirus vs RBD)🤔?? 4/
biorxiv.org/content/10.110…
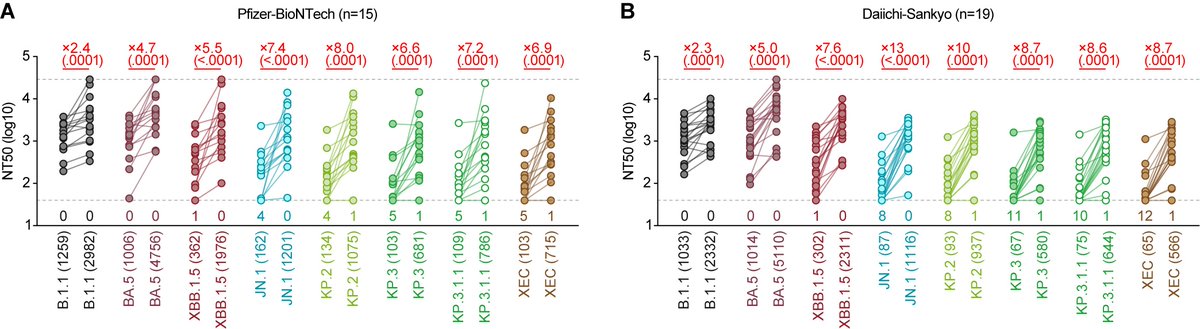
Immune resistance:
In all cases tested, the 50% neutralization titers (NT50s) of XEC and LP.8.1 were comparable. So... why is the Re of LP.8.1 higher than that of XEC...🤔? 5/
In all cases tested, the 50% neutralization titers (NT50s) of XEC and LP.8.1 were comparable. So... why is the Re of LP.8.1 higher than that of XEC...🤔? 5/
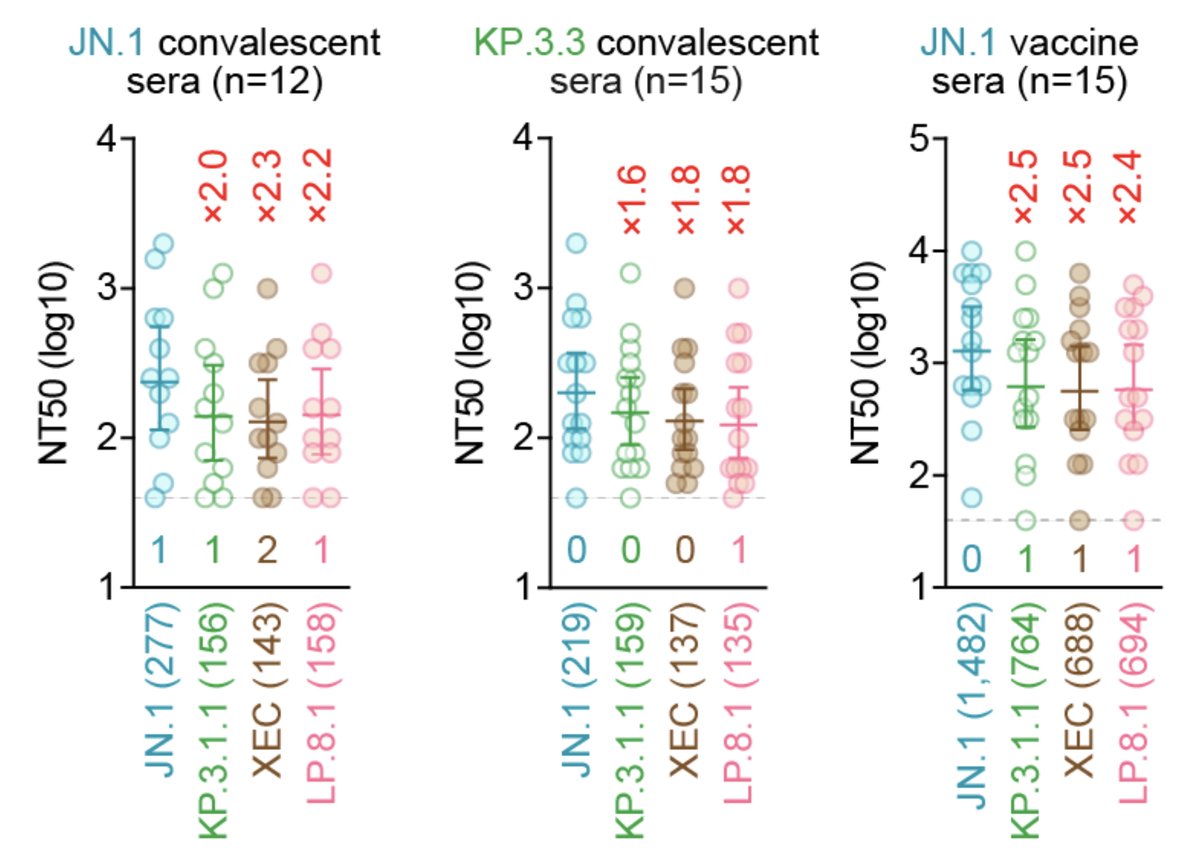
The advantage of LP.8.1 over XEC may depend on the immune background and/or the history of SARS-CoV-2 infection/vaccination of the population in each country? Anyways both live-virus and real-world data from more countries would be needed to better understand of LP.8.1... 6/
Over the next few months, pharmaceutical companies will be considering the vaccine variant for fall/winter 2025. It would be particularly important to take into account our data suggesting that LP.8.1 may NOT be dominant globally when considering the vaccine candidate. 7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh