We received an (indirect) response from UNC regarding our preprint outlining evidence for a potential lab leak of SARS2 at UNC
UNC asked GISAID to tell us to remove our preprint
We did this, but have now put up an updated version 🧵
zenodo.org/records/151721…
UNC asked GISAID to tell us to remove our preprint
We did this, but have now put up an updated version 🧵
zenodo.org/records/151721…
2/ @quay_dr uploaded our original preprint on March 26
We identified 7 'frozen' SARS2 sequences from UNC, that were basal to other sequences generated at the same time. These could represent early SARS2 strains that had escaped from a research facility
We identified 7 'frozen' SARS2 sequences from UNC, that were basal to other sequences generated at the same time. These could represent early SARS2 strains that had escaped from a research facility
https://x.com/stevenemassey/status/1904619032151810403
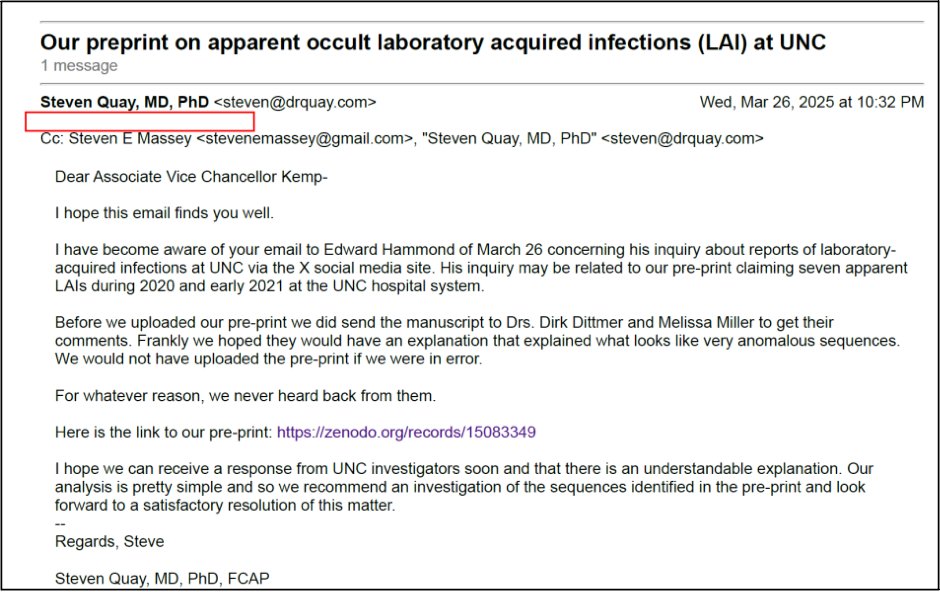
3/ Before uploading the preprint, we sent an email to Dirk Dittmer and Melissa Miller of the Clinical Molecular Microbiology Laboratory, UNC Hospital, regarding the 7 anomalous sequences we had detected from their sequencing facility
However, they did not respond
However, they did not respond

4/ We also sent an email to Dr Derek Kemp, the UNC Associate Vice Chancellor for Campus Safety and Risk Management, alerting him to our preprint and asking for clarification
However, we received no response
However, we received no response
5/ UNC have not contacted my co-author @quay_dr or myself directly
Rather, both UNC and the CDC (!?) contacted GISAID, who then phoned @quay_dr to take down our preprint as it contained a fasta file of the 7 sequences in the preprint supplementary material
Rather, both UNC and the CDC (!?) contacted GISAID, who then phoned @quay_dr to take down our preprint as it contained a fasta file of the 7 sequences in the preprint supplementary material
6/ The reason we included the fasta sequences was for reproducibility and transparency
However, GISAID objected that this violated their user agreement and requested that we use a hyperlink to direct investigators to the fasta data, which we were happy to do
However, GISAID objected that this violated their user agreement and requested that we use a hyperlink to direct investigators to the fasta data, which we were happy to do
7/ This is strange behavior from UNC, and the CDC
Why not address the substance of our preprint, that proposes that the 7 sequences indicate a potential lab leak, rather than engaging in vexatious quibbles ?
Why not address the substance of our preprint, that proposes that the 7 sequences indicate a potential lab leak, rather than engaging in vexatious quibbles ?
8/ The role of the CDC is unclear in this. They are supposed to be an independent agency, why are they lobbying to have UNC-generated sequence data removed from a preprint authored by decentralized investigators ?
9/ One would expect that if an institution is alerted to information that indicates a potential lab leak from one of their facilities, it should address those concerns
This has not happened, rather UNC engaged in evasive tactics, indicating a flaw in biosafety procedures
This has not happened, rather UNC engaged in evasive tactics, indicating a flaw in biosafety procedures
10/ If this is the response from institutions engaged in dangerous pathogen research to inconvenient sequence data, how can we be assured that in future such sequences are not simply discarded by the sequencing facility, rather than making them public ?
11/ There is currently no proposal to address this key problem in genomic surveillance, that of conflict of interest between sequencing centers and the public
One solution would be to use blockchain to ensure the chain of custody is not violated and information not censored
One solution would be to use blockchain to ensure the chain of custody is not violated and information not censored
12/ If sequences can be judged to be 'noisy' or 'artefactual', and removed as a consequence, we need to be sure that this is not used as an excuse to censor bona fide 'frozen' genome sequences, which are one of the red flags indicating a potential leak
mdpi.com/2076-2607/12/1…
mdpi.com/2076-2607/12/1…
13/ The other red flag is the institutional response to public concerns of a potential pathogen leak
In the example reported here, the response of UNC is more consistent with a leak
In the example reported here, the response of UNC is more consistent with a leak
14/ The nature of anomalous virus sequences can often only be clarified with the cooperation of the originating institution. If that cooperation is not forthcoming then a question mark remains
Any indication of a potential leak should trigger an automatic investigation
Any indication of a potential leak should trigger an automatic investigation
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh










