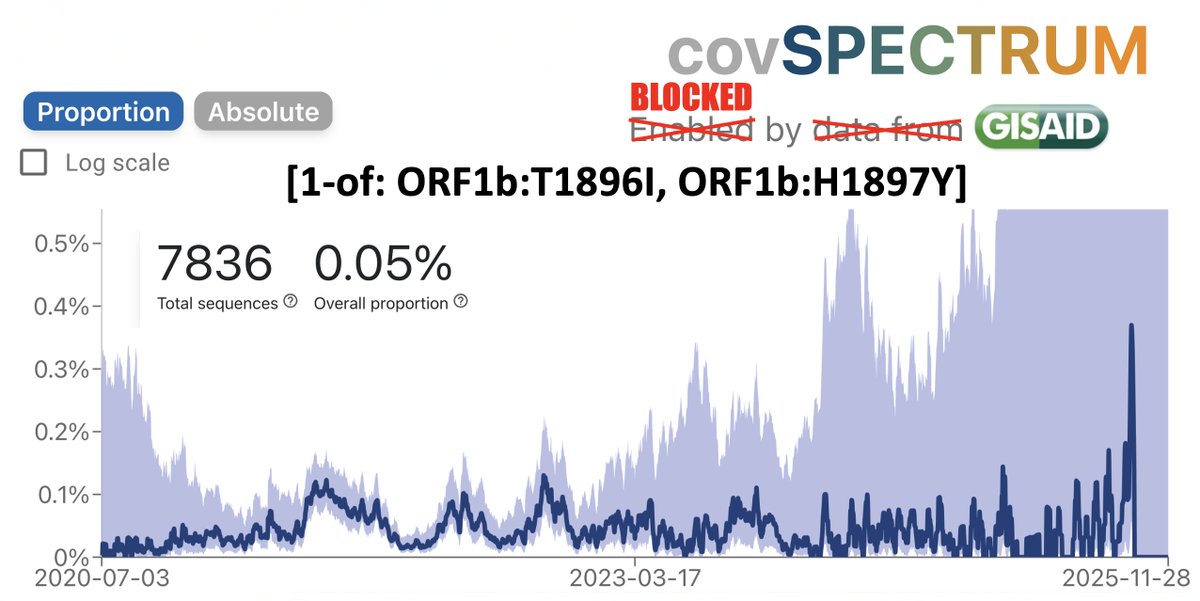
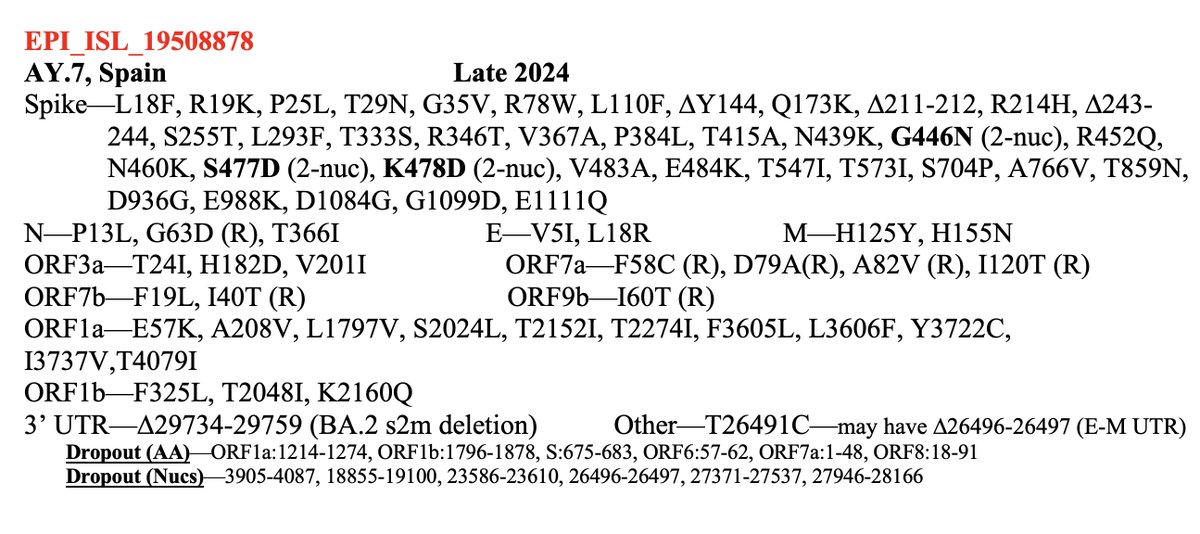
Incredible how quickly @yunlong_cao & co provide us w/info on the latest emerging SARS-CoV-2 variants.
Already, we have great data on BA.3.2 (the divergent saltation lineage detected in South Africa & the Netherlands & NB.1.8.1, an emerging contender for global dominance. 1/9

Already, we have great data on BA.3.2 (the divergent saltation lineage detected in South Africa & the Netherlands & NB.1.8.1, an emerging contender for global dominance. 1/9


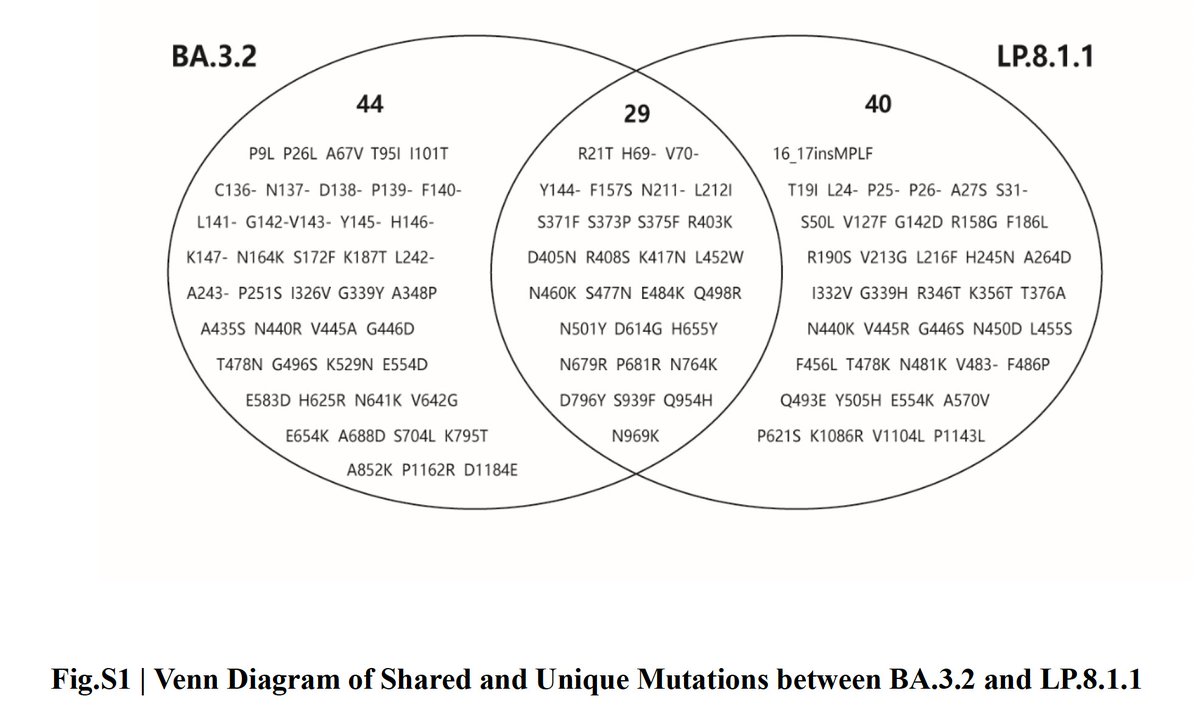
BA.3.2 is a clear outlier on the antigenic cartography map—as expected given the enormous differences between its spike protein & every other circulating variant. 2/9

https://x.com/LongDesertTrain/status/1899647059872850369

It's unsurprising, therefore, that BA.3.2 evades antibodies from human sera more effectively than any other variant, though the degree of its superiority is striking. 3/9
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
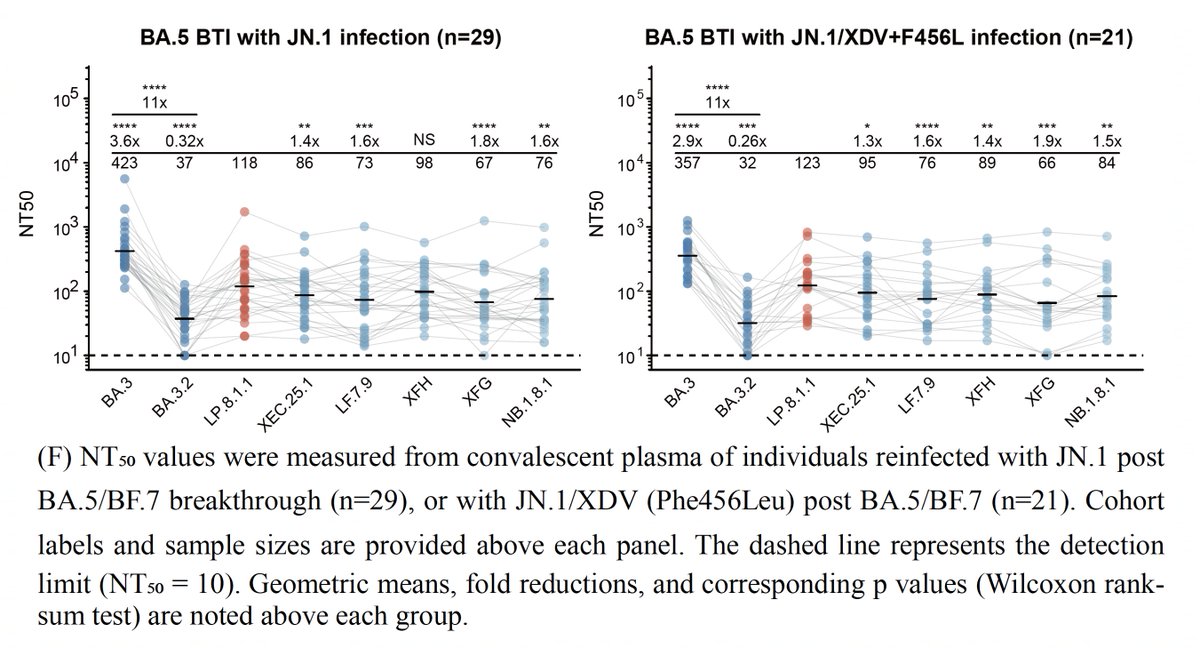
Then why hasn't BA.3.2 grown and spread very much? Its ACE2 binding is quite weak, & most of this is due to its tendency to be in the "closed" RBD conformation, which helps evade antibodies but reduces ACE2 interaction & infectivity, a topic @jbloom_lab has explored. 4/9 
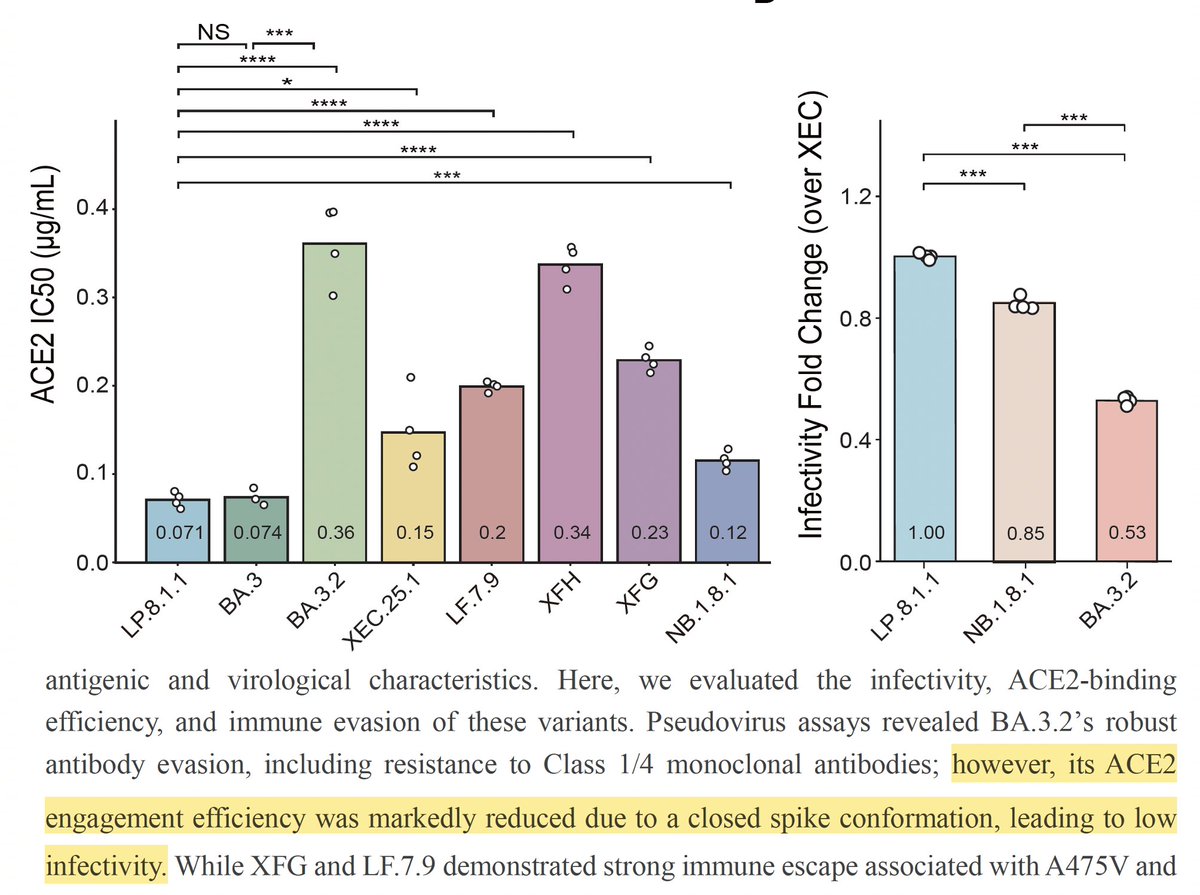
NB.1.8.1 and LP.8.1.1, which seem the two most likely contenders for future dominance, have quite similar spikes, though NB.1.8.1 exhibits slightly greater antibody evasion here. However, the sera was from patients in China, where NB.1.8.1 has had its greatest success. 5/9 
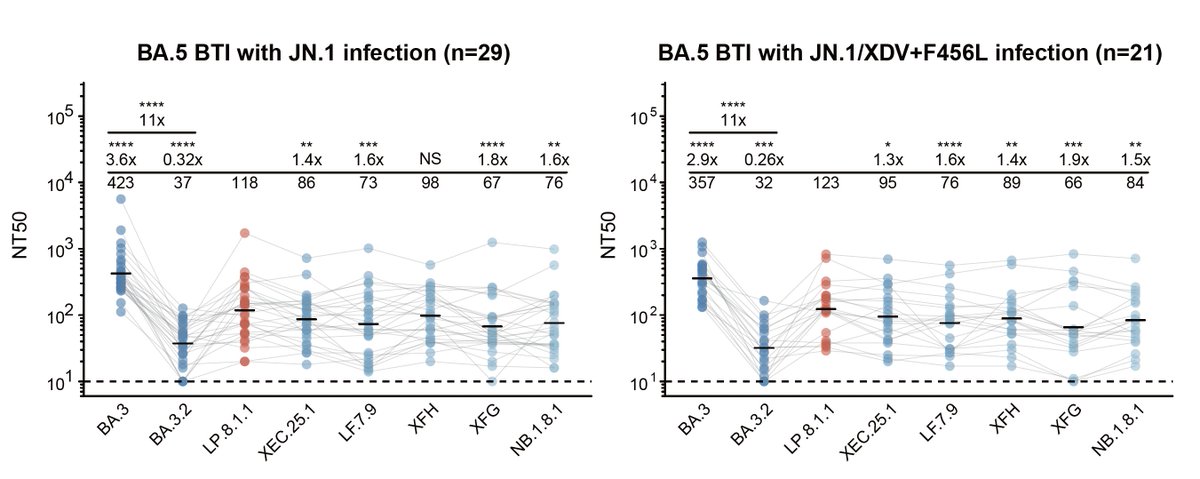
The population immunity environment is still, IMO, different in China (& to some extent in Japan, AUS, & NZ) than elsewhere, so whether this translates into rapid global growth is still an open question. However, NB.1.8.1 is starting to appear & grow a bit in USA & Canada. 6/9
See, for example, @SolidEvidence's excellent wastewater surveillance tool, Lungfish. While only at 1-2%, NB.1.8.1 mutations are growing. Some of this may be due to exports from China (which may be undergoing a Covid wave), though how much is uncertain. 7/ lungfish-science.github.io/wastewater-das…
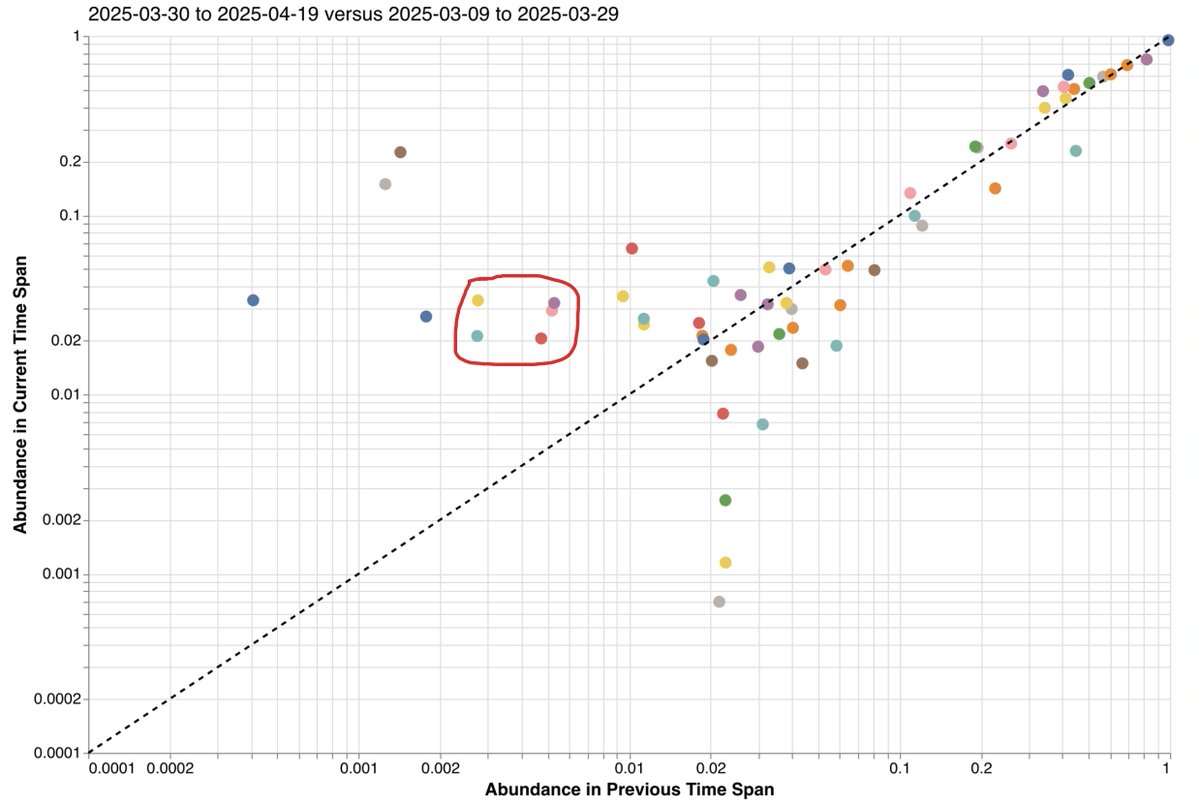
Regardless of whether LP.8.1.1 or NB.1.8.1 wins out, the spike changes in both are modest compared to the variants of the past 8-10 months, so I don't think either is likely to have a significant effect on overall case levels.
8/9
8/9
BA.3.2, on the other hand, has characteristics of a variant that could drive a global wave, but its closed spike & weak ACE2 interaction mean that it would need to acquire additional adaptations in order to be widely successful. 9/end 
• • •
Missing some Tweet in this thread? You can try to
force a refresh