🔬Simplified breakdown of the Decode ME results:
DecodeME identifies 8 gene regions linking immune response, mitochondrial energy control, and brain-cell signalling to ME/CFS
Genomic evidence the disease is biological.
Let’s breakdown everything in depth 🧵
DecodeME identifies 8 gene regions linking immune response, mitochondrial energy control, and brain-cell signalling to ME/CFS
Genomic evidence the disease is biological.
Let’s breakdown everything in depth 🧵

Study Cohort:
15,579 people with doctor-diagnosed ME/CFS + 259,909 UK Biobank controls (no ME/CFS).
85 % were women, average age ≈ 52 y.
15,579 people with doctor-diagnosed ME/CFS + 259,909 UK Biobank controls (no ME/CFS).
85 % were women, average age ≈ 52 y.
How much is genetic?
Common SNPs explain = 9.5 % of overall ME/CFS risk (heritability on the liability scale).
For comparison:
- asthma = 10 %
- arthritis = 12 %
- type 2 diabetes = 13%
So ME/CFS is typical for complex diseases when you look only at common variants.
Common SNPs explain = 9.5 % of overall ME/CFS risk (heritability on the liability scale).
For comparison:
- asthma = 10 %
- arthritis = 12 %
- type 2 diabetes = 13%
So ME/CFS is typical for complex diseases when you look only at common variants.
Key finding:
8 DNA regions change risk a little (odds-ratios ~1.08 ↑ or 0.93 ↓).
OR 1.08 = 8 % higher odds of developing ME
OR 0.93 = 7 % lower odds of developing ME
Multiple genes stack up to increase or lower risk.
8 DNA regions change risk a little (odds-ratios ~1.08 ↑ or 0.93 ↓).
OR 1.08 = 8 % higher odds of developing ME
OR 0.93 = 7 % lower odds of developing ME
Multiple genes stack up to increase or lower risk.
Main genes & plain meanings:
- RABGAP1L (helps cells expel germs)
- BTN2A2 (activates a special T-cell)
- FBXL4 (keeps mitochondria healthy [energy])
- SUDS3 (controls brain immune cells)
- RABGAP1L (helps cells expel germs)
- BTN2A2 (activates a special T-cell)
- FBXL4 (keeps mitochondria healthy [energy])
- SUDS3 (controls brain immune cells)
- OLFM4 (tones down neutrophil bug-killing)
- CCPG1 (cleans stressed ER parts)
- CA10 (shapes nerve-to-nerve contacts)
- ARFGEF2 / CSE1L (manage TNF-α, an inflammation signal)
- CCPG1 (cleans stressed ER parts)
- CA10 (shapes nerve-to-nerve contacts)
- ARFGEF2 / CSE1L (manage TNF-α, an inflammation signal)
Where do these genes matter most?
A tool called MAGMA (it groups DNA signals by gene and checks which tissues use those genes) shows they’re used most in the brain. So the genetic clues link ME/CFS to the nervous system as well as the immune system.
A tool called MAGMA (it groups DNA signals by gene and checks which tissues use those genes) shows they’re used most in the brain. So the genetic clues link ME/CFS to the nervous system as well as the immune system.
Does infection matter?
Yes. In people whose illness began after an infection, the OLFM4 signal is much stronger; it’s absent in non-infection cases.
Yes. In people whose illness began after an infection, the OLFM4 signal is much stronger; it’s absent in non-infection cases.
Male vs female DNA effects?
Variants act equally in men and women; male-only analysis lacked power but key female hits (CA10, ARFGEF2) still showed the same direction.
Variants act equally in men and women; male-only analysis lacked power but key female hits (CA10, ARFGEF2) still showed the same direction.
Immune insights:
HLA allele DQA1*05:01 was slightly protective (less common in patients). HLA genes help immune cells recognise threats.
HLA allele DQA1*05:01 was slightly protective (less common in patients). HLA genes help immune cells recognise threats.
Overlap with other diseases?
- The CA10 region is shared with multisite chronic pain (high probability it’s the same causal SNP).
- None of the eight regions share causal SNPs with depression or anxiety studies.
- The CA10 region is shared with multisite chronic pain (high probability it’s the same causal SNP).
- None of the eight regions share causal SNPs with depression or anxiety studies.
So will this help identify treatments?
When a disease-linked gene pinpoints a process (e.g., TNF-α release or mitochondrial upkeep) drug projects aimed at that process have higher success rate of projects without genetic support.
When a disease-linked gene pinpoints a process (e.g., TNF-α release or mitochondrial upkeep) drug projects aimed at that process have higher success rate of projects without genetic support.
Example 1 - Inflammation angle
DecodeME noted a region with the genes ARFGEF2 / CSE1L that regulate how cells package and release TNF-α, a key inflammatory signal.
Existing anti-TNF drugs (used in rheumatoid arthritis & Crohn’s) could now be tested for ME/CFS.
DecodeME noted a region with the genes ARFGEF2 / CSE1L that regulate how cells package and release TNF-α, a key inflammatory signal.
Existing anti-TNF drugs (used in rheumatoid arthritis & Crohn’s) could now be tested for ME/CFS.
Example 2 - Nerve-signalling angle
Another hit, CA10, shares the same causal variant with multisite chronic pain.
CA10 affects how nerve cells talk to each other. Compounds that fine-tune this synaptic pathway (already explored for pain) are now candidates to check in ME/CFS.
Another hit, CA10, shares the same causal variant with multisite chronic pain.
CA10 affects how nerve cells talk to each other. Compounds that fine-tune this synaptic pathway (already explored for pain) are now candidates to check in ME/CFS.
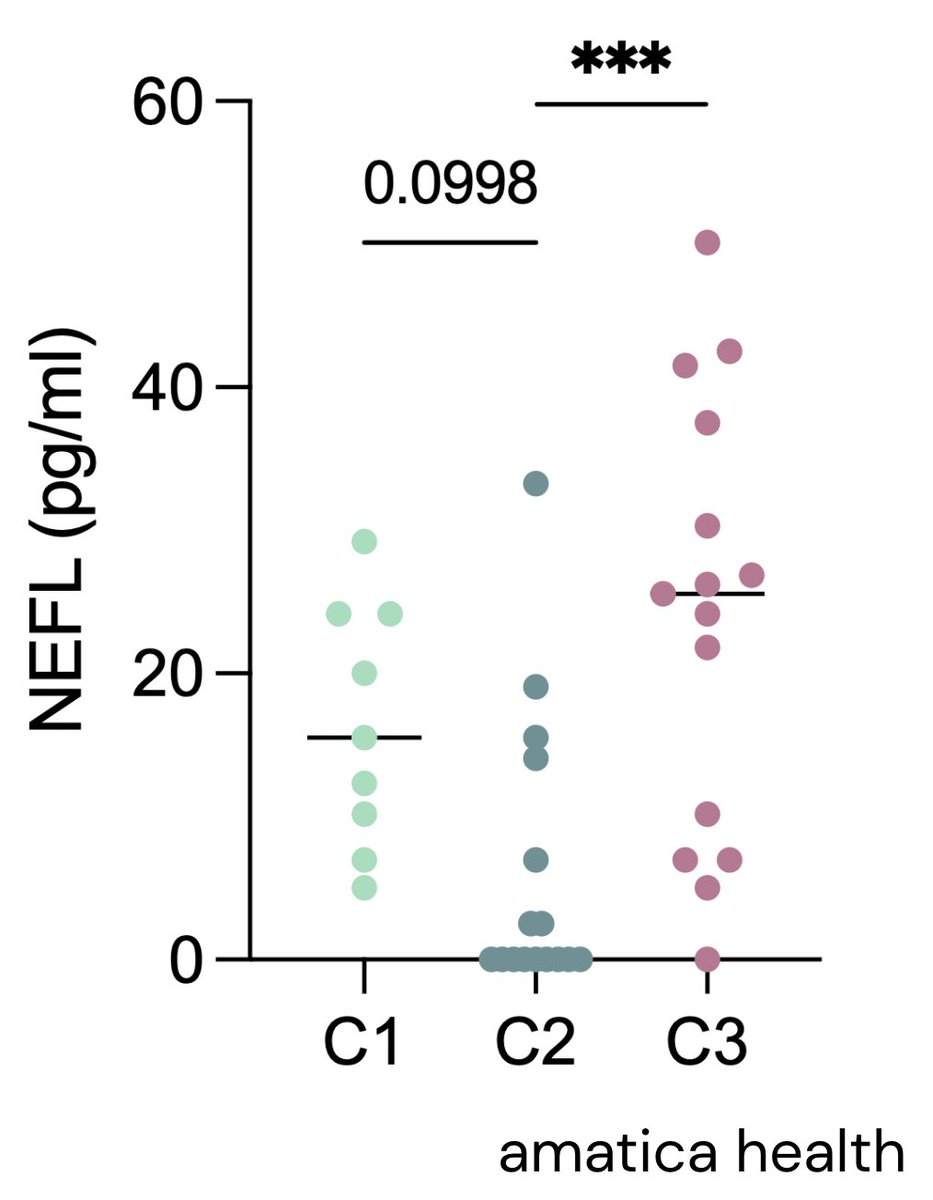
Will we be able to expand on insights with our new @amaticahealth RNA seq test?
If a person carries a “risk” or “protective” version of immune-expressed genes like RABGAP1L, BTN2A2, OLFM4, ARFGEF2, CSE1L we can see if their RNA level go up or down.
If a person carries a “risk” or “protective” version of immune-expressed genes like RABGAP1L, BTN2A2, OLFM4, ARFGEF2, CSE1L we can see if their RNA level go up or down.
This allows us to see how the generic variant is impacting the functioning of the system.
Example:
If a risk DNA drops RABGAP1L RNA (weaker bug-clearing) or boosts OLFM4 RNA (stronger neutrophil brake), the up/down shift shows whether that variant turns immune defences down or inflammation up
If a risk DNA drops RABGAP1L RNA (weaker bug-clearing) or boosts OLFM4 RNA (stronger neutrophil brake), the up/down shift shows whether that variant turns immune defences down or inflammation up
So overall very much what we expected from the study.
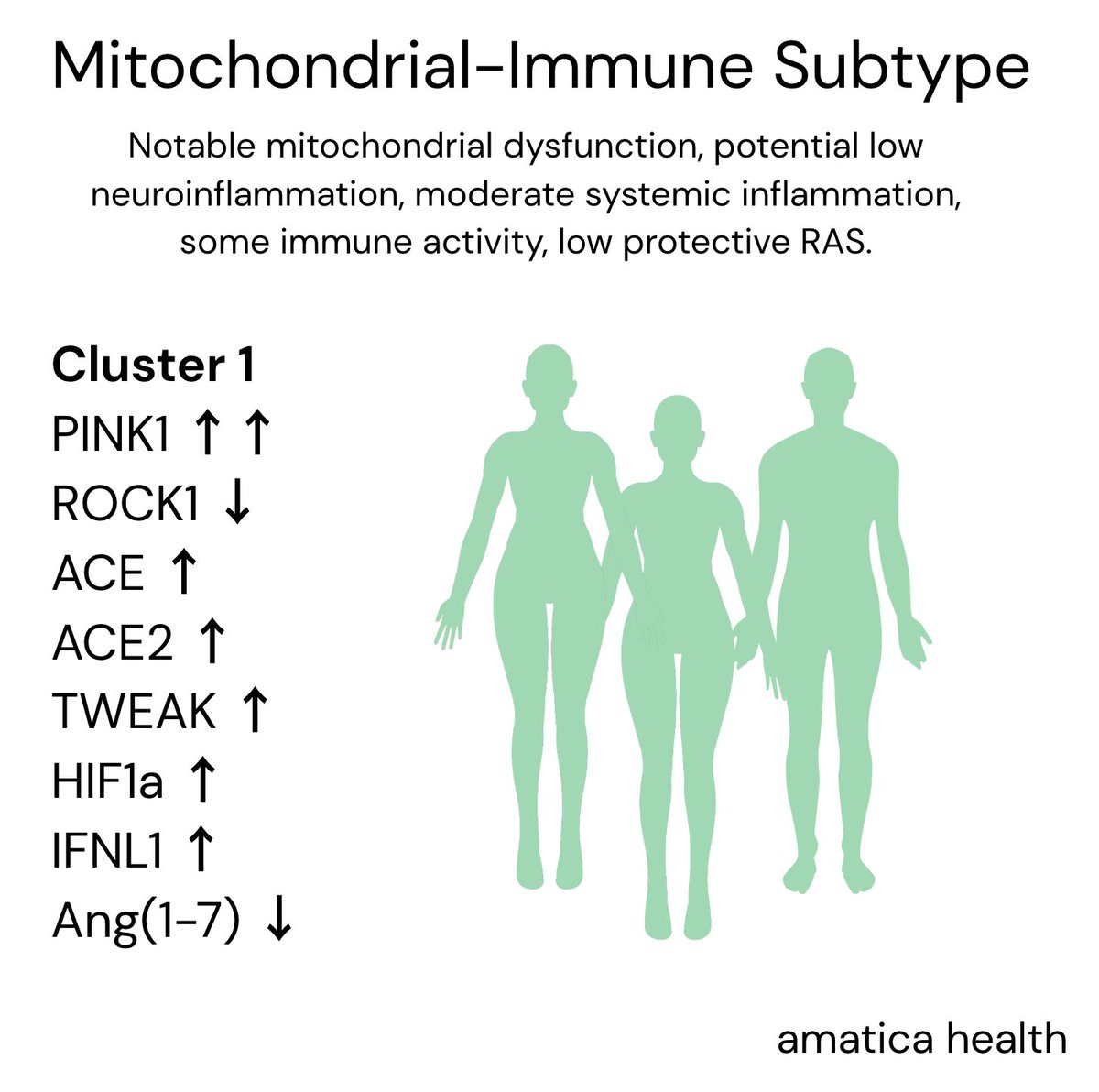
Risk factor genes that relate to immune system, mitochondria, and nervous system function.
The necessary next steps now are to determine if these alterations cause functional changes that can drive the disease.
Risk factor genes that relate to immune system, mitochondria, and nervous system function.
The necessary next steps now are to determine if these alterations cause functional changes that can drive the disease.
Or are they simply just a ‘trigger’ risk.
I will come back to the Decode ME findings in a few months when we have our RNA data to see if we can confirm any changes in gene expression within these similar systems (TNF-a signalling etc)
And track against disease profile.
I will come back to the Decode ME findings in a few months when we have our RNA data to see if we can confirm any changes in gene expression within these similar systems (TNF-a signalling etc)
And track against disease profile.
I will also do some more breakdowns on the exact genes and their prevalence in other diseases, if known, over the next few weeks.
• • •
Missing some Tweet in this thread? You can try to
force a refresh