It’s been 2 years since BA.2.86 first appeared (and I’m give the variant update to SAVE on Monday), so I thought I would do a little summary about this era of SARS-CoV-2 evolution.
1/
1/

SARS-CoV-2 lineages come up with new constellations of mutations in 3 main ways.
1. Sequential acquisition of mutations during normal circulation.
2. Recombination.
3. Sweeping new lineages (almost certainly from persistent infections).
2/
1. Sequential acquisition of mutations during normal circulation.
2. Recombination.
3. Sweeping new lineages (almost certainly from persistent infections).
2/
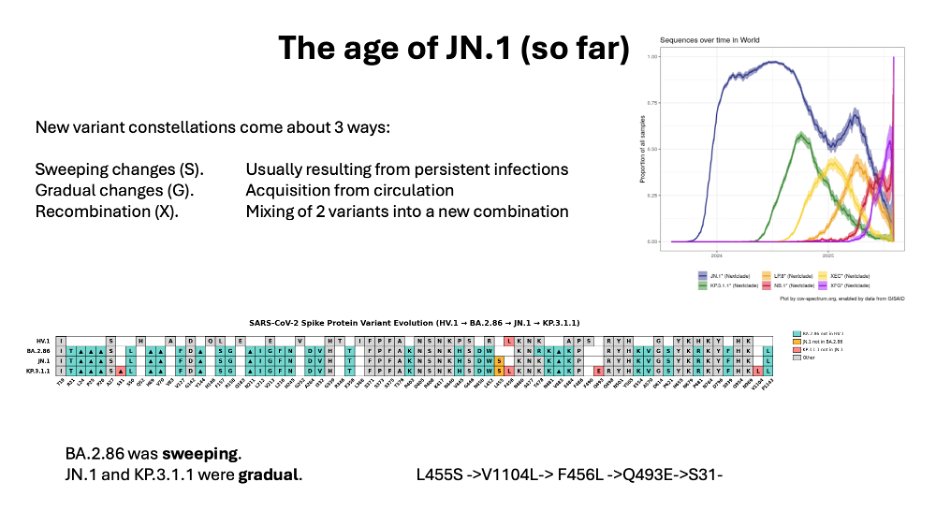
BA.2.86 was one of the sweeping changes. When it first appeared in Israel I thought it was a persistent infection, most of which never spread. Then it appeared in Denmark too.
3/
3/
https://x.com/SolidEvidence/status/1691070379584815104
A few days later it showed up on another continent.
4/
4/
https://x.com/SolidEvidence/status/1692522623169655011
However, our immunity was holding up. There was still a class of antibodies that did a pretty good job of neutralizing BA.2.86.
However, a single AA variation changed that.
The new lineage was designated JN.1
5/
However, a single AA variation changed that.
The new lineage was designated JN.1
5/
https://x.com/SolidEvidence/status/1692522623169655011
JN.1 displaced all other lineages in the world within a few months.
There was a lot of convergent evolution in the JN.1 offspring (+F456L), but no single lineage became dominant until KP.3.1.1 about 6 months later.
6/
There was a lot of convergent evolution in the JN.1 offspring (+F456L), but no single lineage became dominant until KP.3.1.1 about 6 months later.
6/
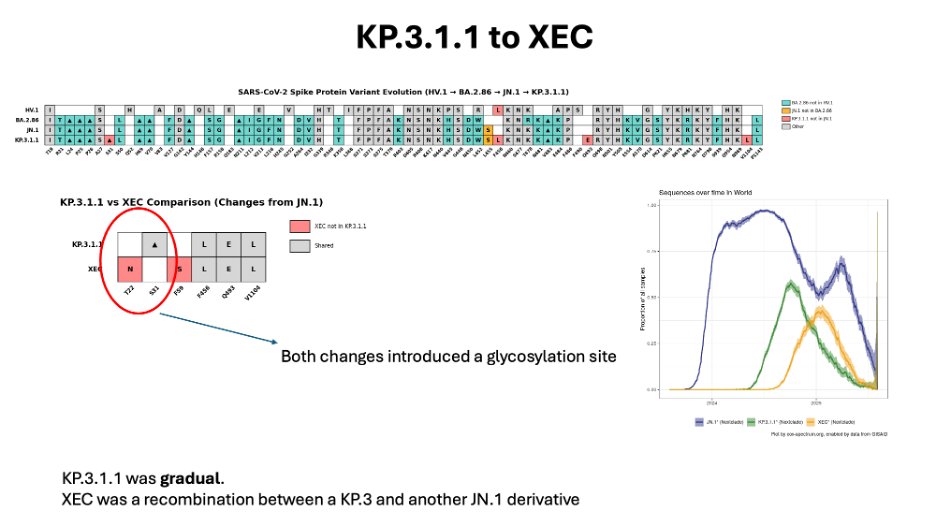
After KP.3.1.1 can the recombinant XEC.
XEC was a recombination of KP.3.1.1 with another JN.1, so KP.3.1.1. and XEC were very similar.
XEC slowly won out over KP.3.1.1, but never became world dominant.
7/
XEC was a recombination of KP.3.1.1 with another JN.1, so KP.3.1.1. and XEC were very similar.
XEC slowly won out over KP.3.1.1, but never became world dominant.
7/
Next came LP.8.1, which was another JN.1 lineage that had been gradually evolving in SE Asia. It displaced XEC, but also did not become world dominant.
8/
8/
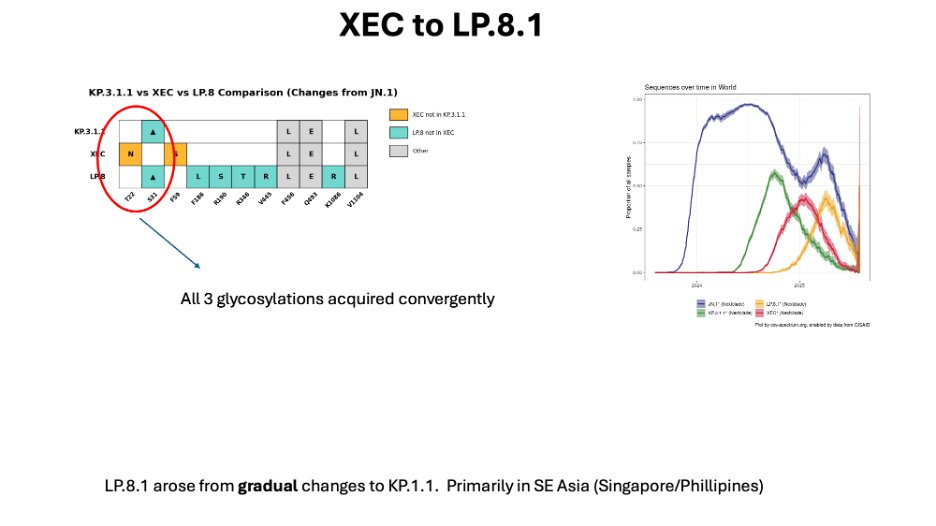
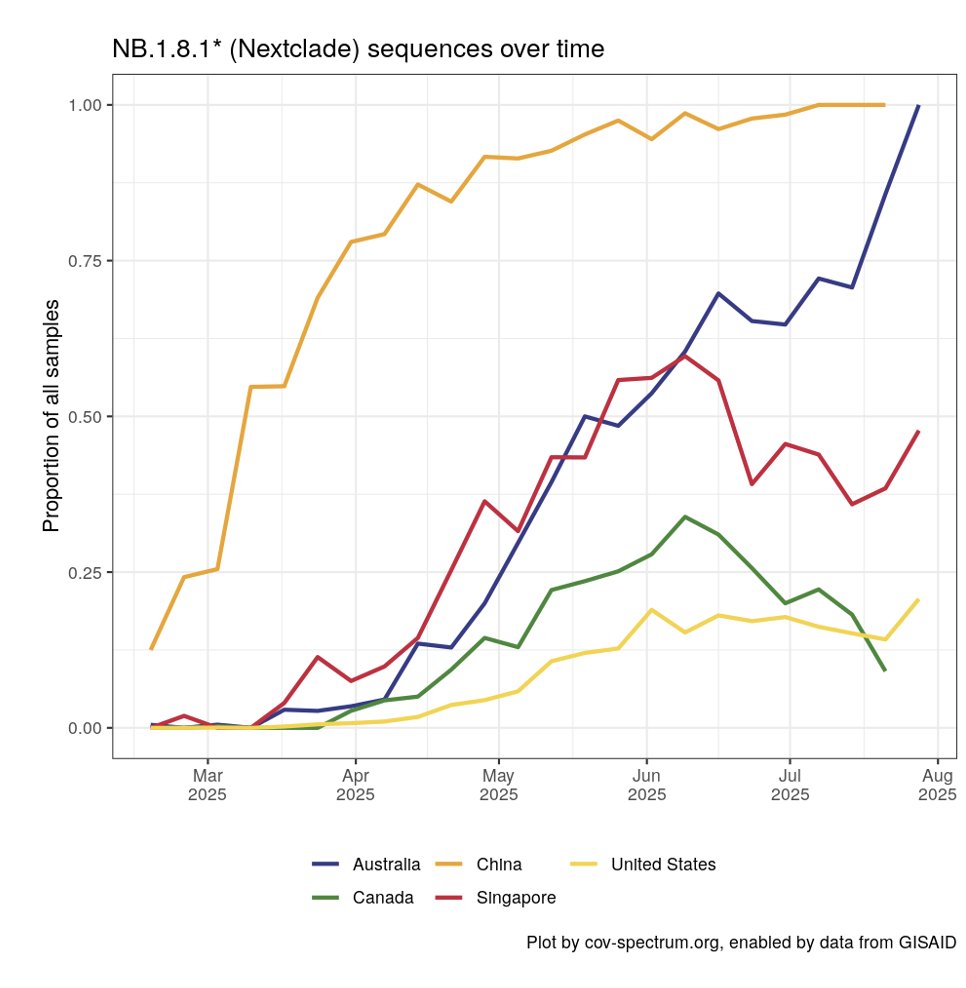
Next came NB.1.8.1, which was derived from a recombinant of a JN.1 with an even older XBB recombinant.
Fun fact, most of the current NB.1.8.1 genome is derived from XBB and not JN.1.
9/
Fun fact, most of the current NB.1.8.1 genome is derived from XBB and not JN.1.
9/
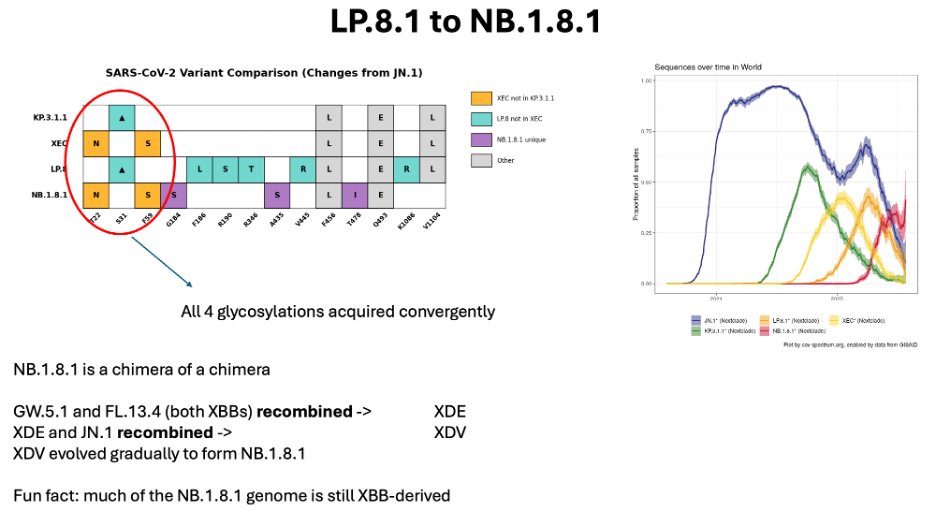
Finally, that brings us to XFG. XFG is a recombinant between LP.8.1 and another JN.1. XFG expanded more rapidly than most of the others, and is the first lineage to reach world dominance (it seems) in a year. 
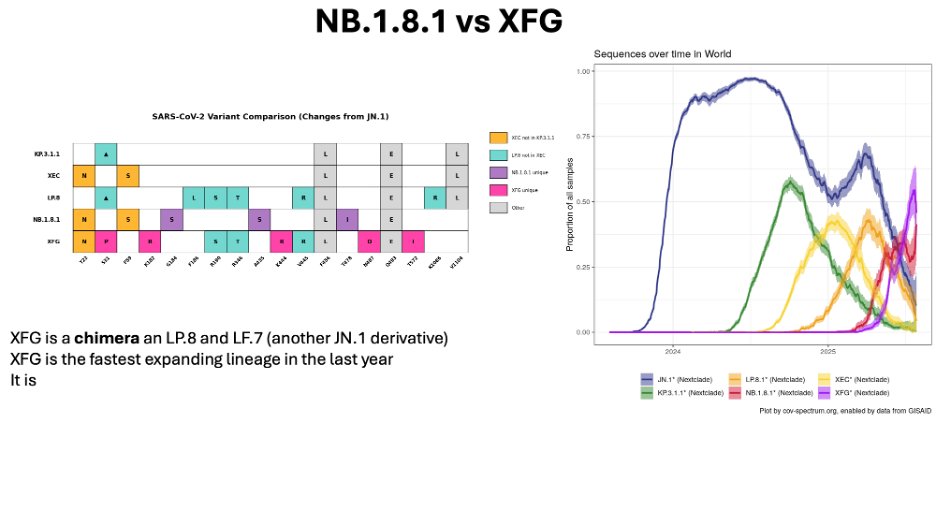
This is our US wastewater readout for XFG. Steadily growing, and appears to be over 60% of the US sequences. There is a corresponding surge (small one) in total virus, which likely is variant driven.
11/
11/
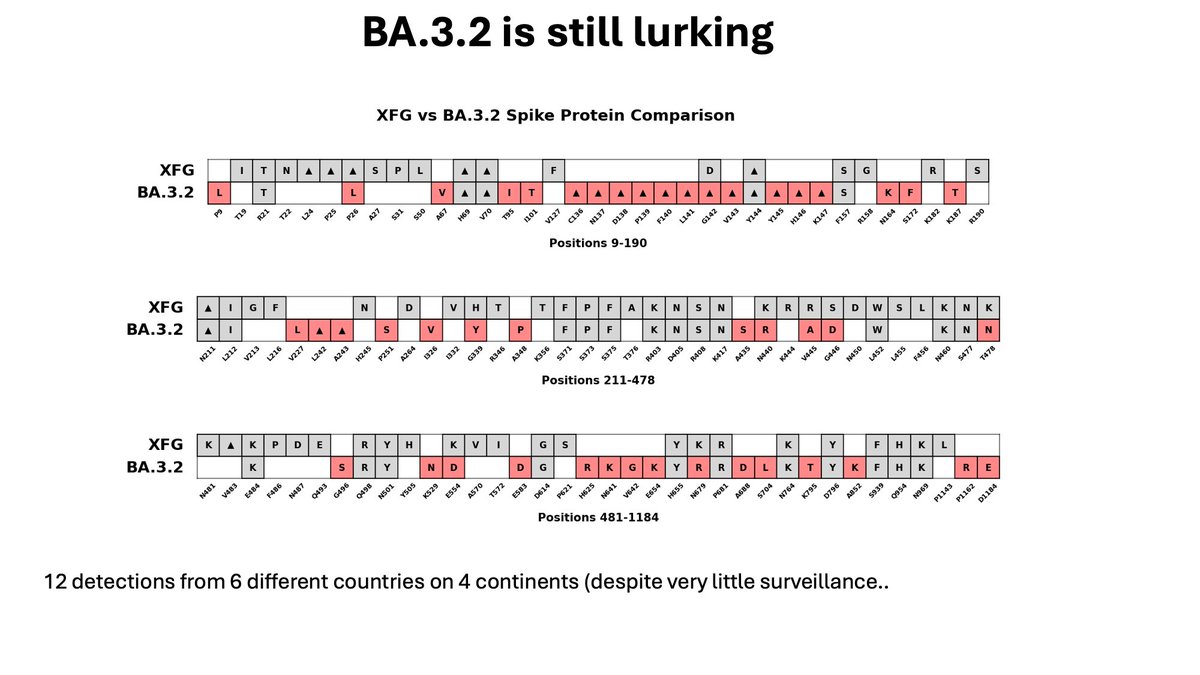
I’m not sure what is next, but we are all still keeping an eye on BA.3.2. This sweeping lineage appeared a few months ago. It hasn’t pulled a ‘JN.1’ yet, but it has appeared on 4 continents.
We’ll be watching.
13/13
We’ll be watching.
13/13
• • •
Missing some Tweet in this thread? You can try to
force a refresh