A fascinating new preprint w/one very unexpected finding suggests, I believe, that a large proportion of Long Covid may be due to chronic infection in a particular bodily niche, which could be crucial for finding effective LC treatments. It requires some explaining. 🧵 1/33 

First, a brief summary of the relevant parts of the preprint. They examined 30 people (from NIH RECOVER cohort) for 6 months after they had Covid, taking detailed blood immunological markers at 3 time points. 20 had Long Covid (PASC), 10 did not (CONV). 2/ biorxiv.org/content/10.110…

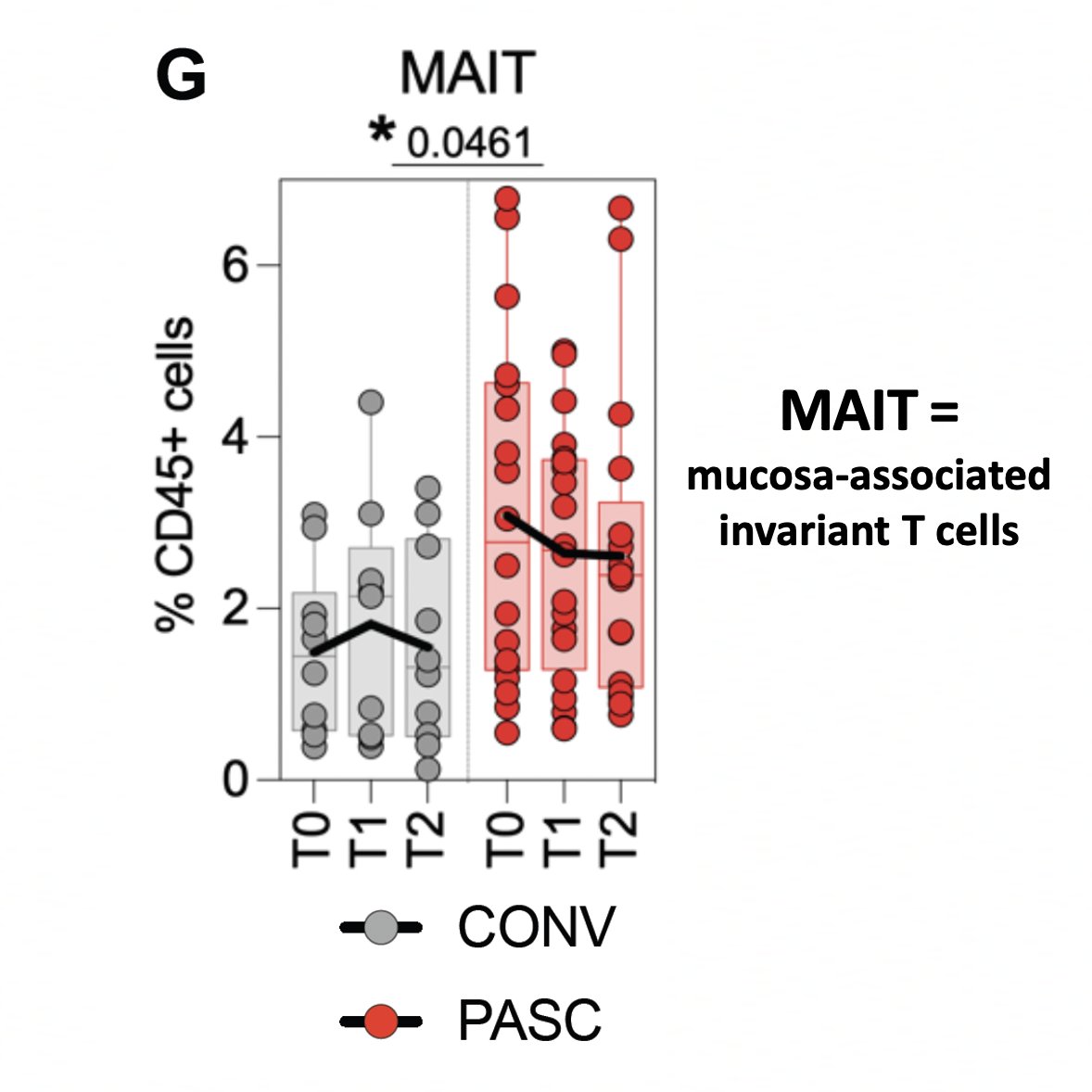
The PASC group showed signs of persistent, pro-inflammatory immune activation over the 6-month time period that suggested ongoing mucosal immune responses, including elevated levels of mucosa-associated invariant T cells (MAIT). 3/ 
Most interesting to me was the peculiar nature of the antibody response in the PASC group. First, unlike in the CONV (control) group, antibody (Ab) levels did not significantly wane over time in the PASC group. High levels of circulating T follicular helper cells (cTFH)… 4/ 
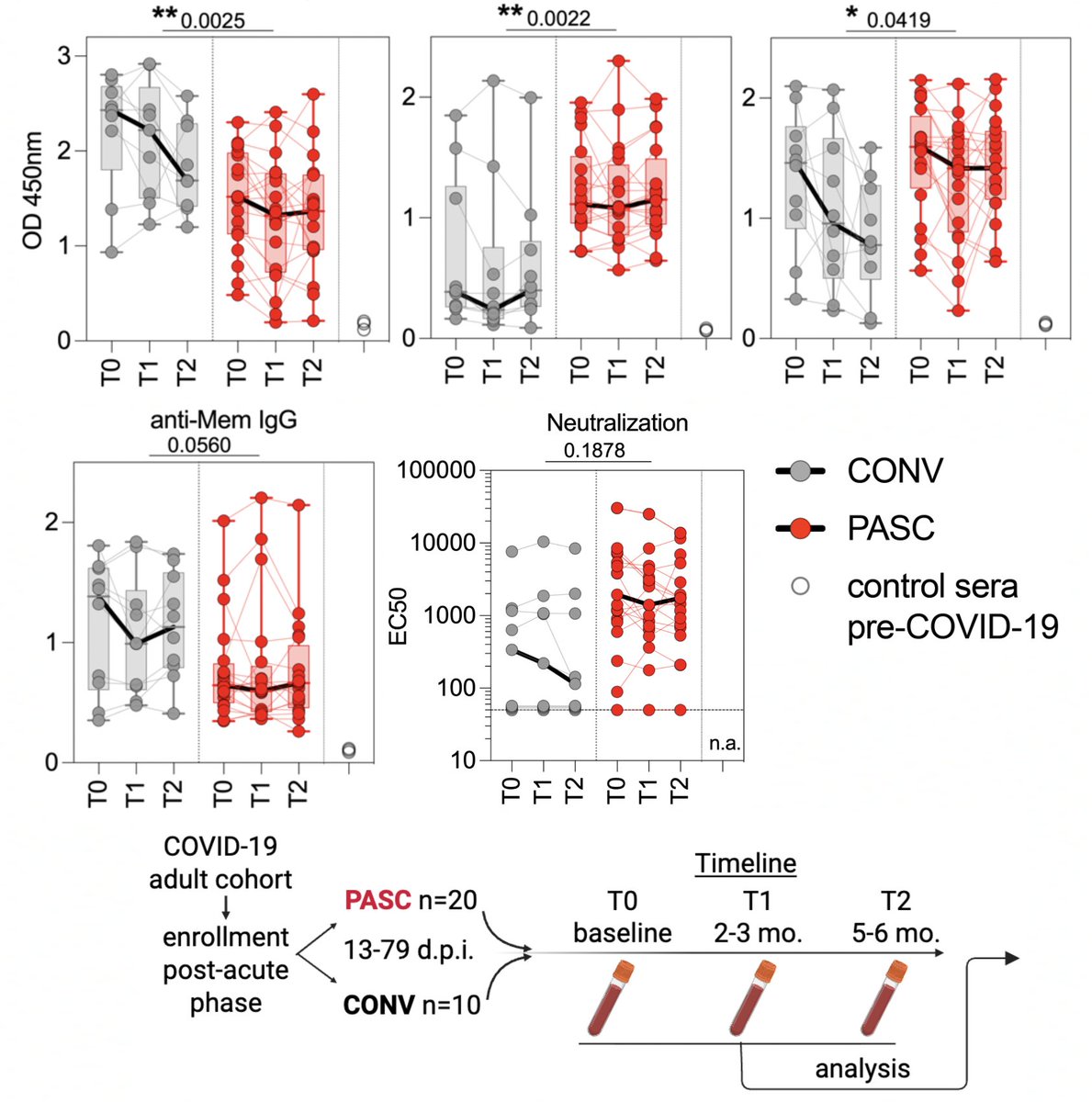
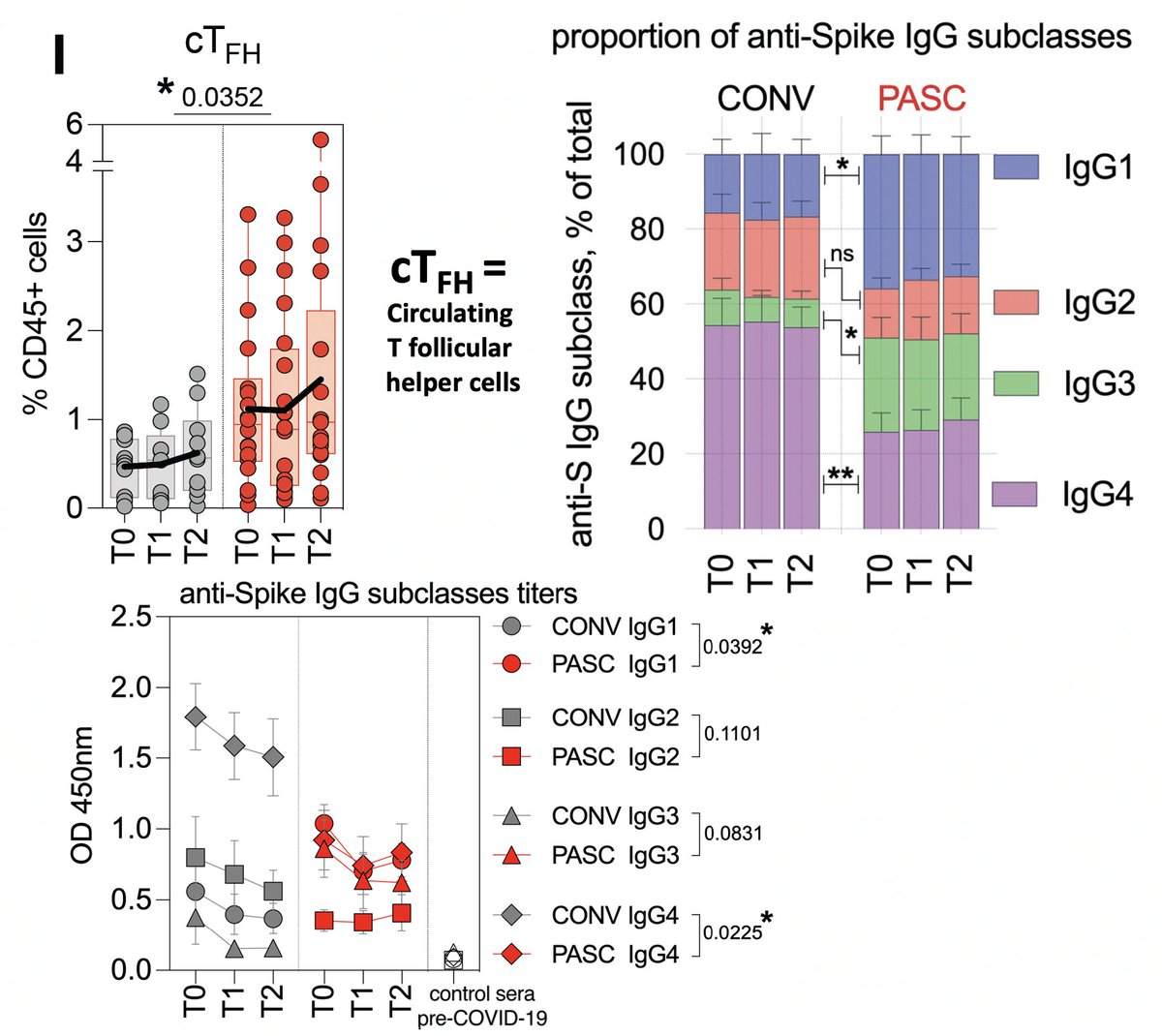
…indicate ongoing germinal center activation (meaning B cells are continually being trained to produce improved, stronger-binding Abs). PASC IgG were skewed toward the more pro-inflammatory IgG1 & IgG3 with much lower IgG4 than CONV patients. 5/ 
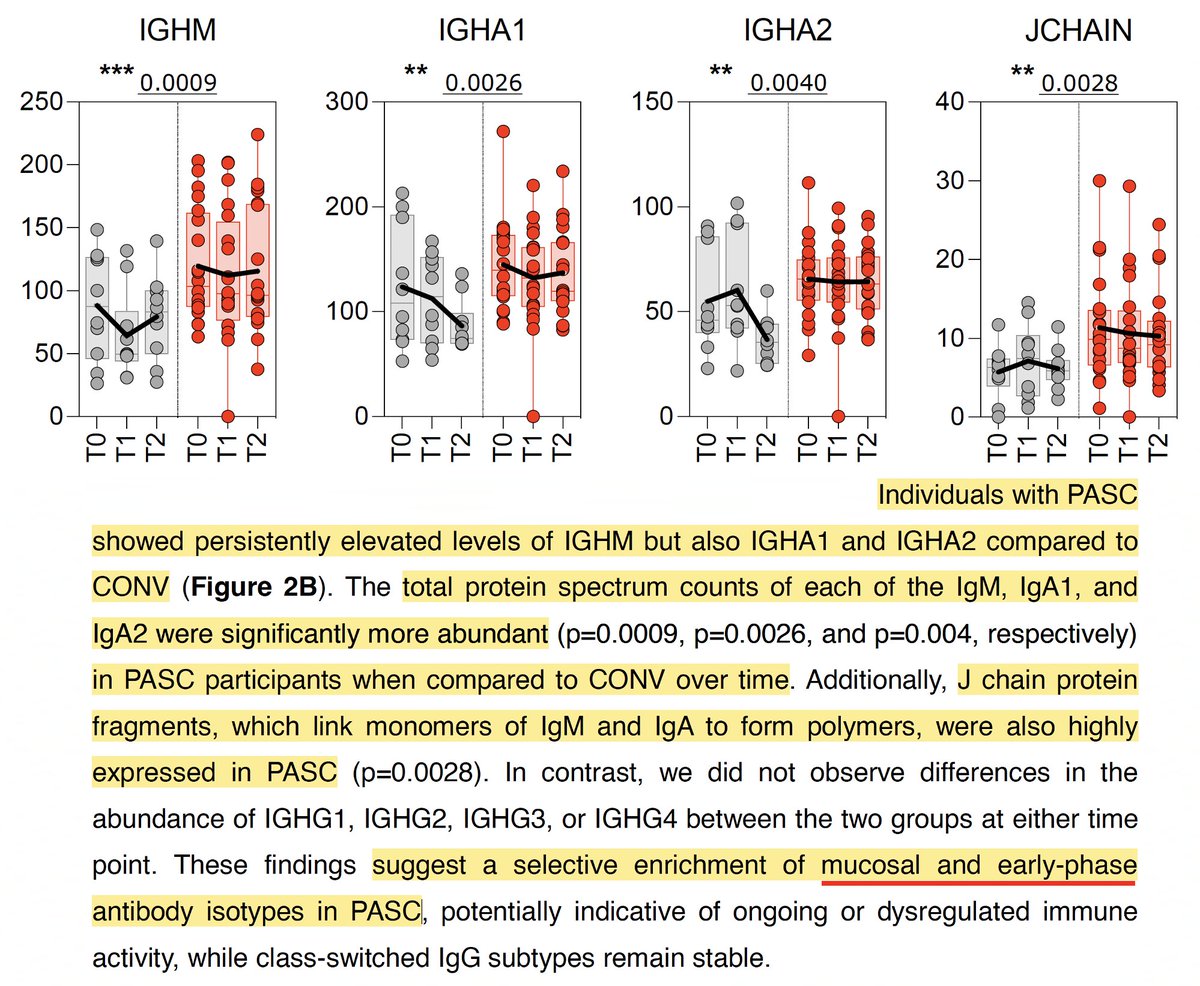
Persistently elevated levels of antibodies typically associated with acute infection, like IgM and J-chain protein fragments, also point toward continual exposure to viral antigens. 6/ 
Most intriguing to me was the antibody targets. Spike-specific Ab titers were much lower in PASC than in the CONV/control group, while Abs targeting envelope (E) were persistently elevated in PASC. 7/ 
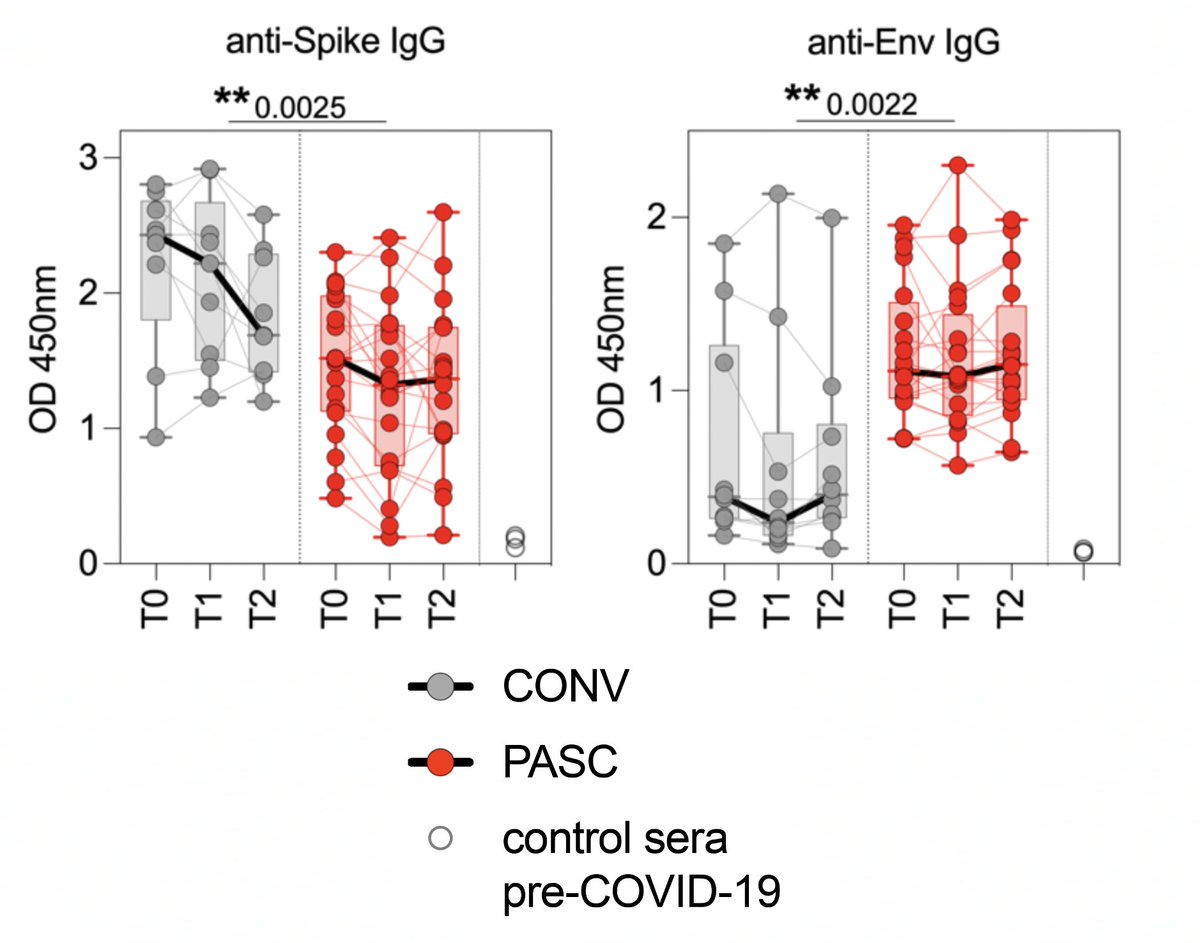
Furthermore, high levels of anti-E antibodies were associated w/higher MAIT counts—which "may reflect persistence of viral antigens in mucosal compartments." 8/ 
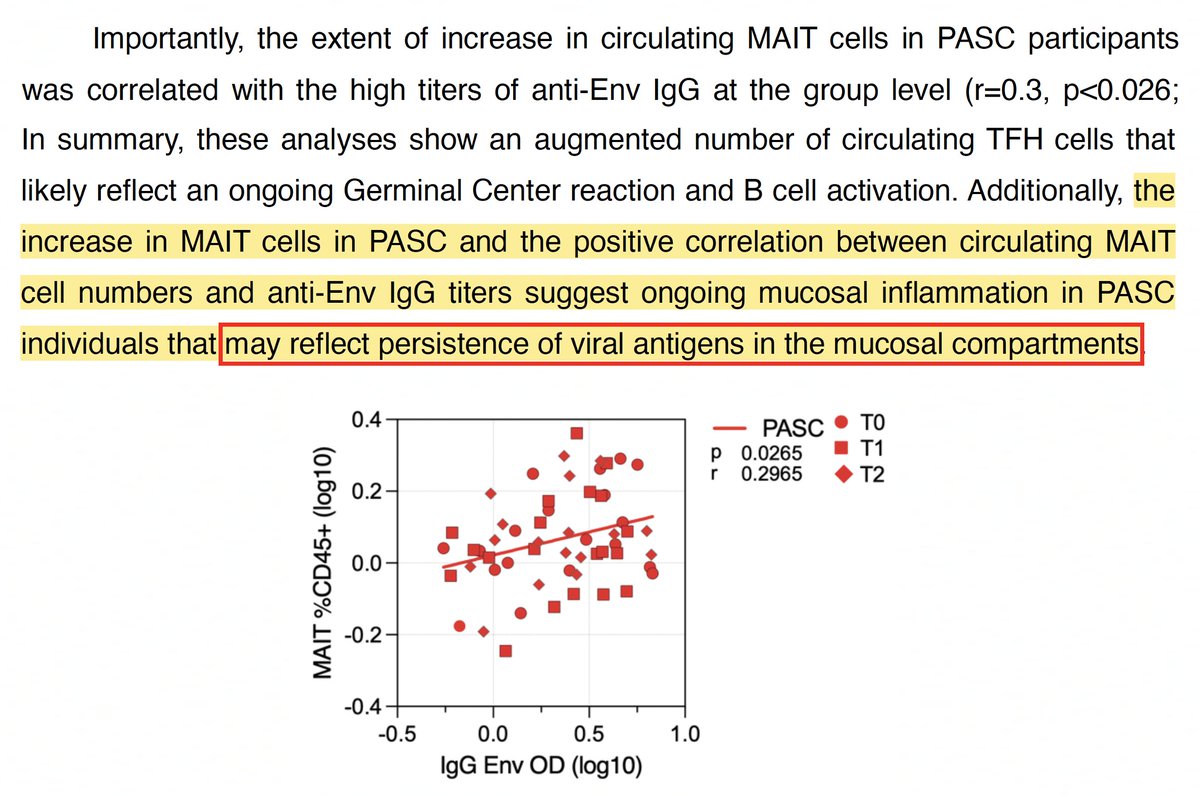
It's all very suggestive of viral persistence, but what to make of the peculiarly high antibodies to E & unusually low antibodies to spike? I don't know the underlying reason for it, but there is a specific category of chronic-infection sequences fits this description. 9/
There are about 10 distinct non-spike mutational patterns that recur again and again in chronic-infection sequences. One is quite different from the others as it involves a large number of regions and is the only pattern to include spike mutations. 10/
Though the full mutational pattern is very diverse, its strongest & most distinctive aspects are three:
#1. Extraordinarily high rate of E mutations
#2. Specific mutations in NSP4
#3. A marked *absence* of spike RBD, NTD, & especially RBM mutations
11/
#1. Extraordinarily high rate of E mutations
#2. Specific mutations in NSP4
#3. A marked *absence* of spike RBD, NTD, & especially RBM mutations
11/
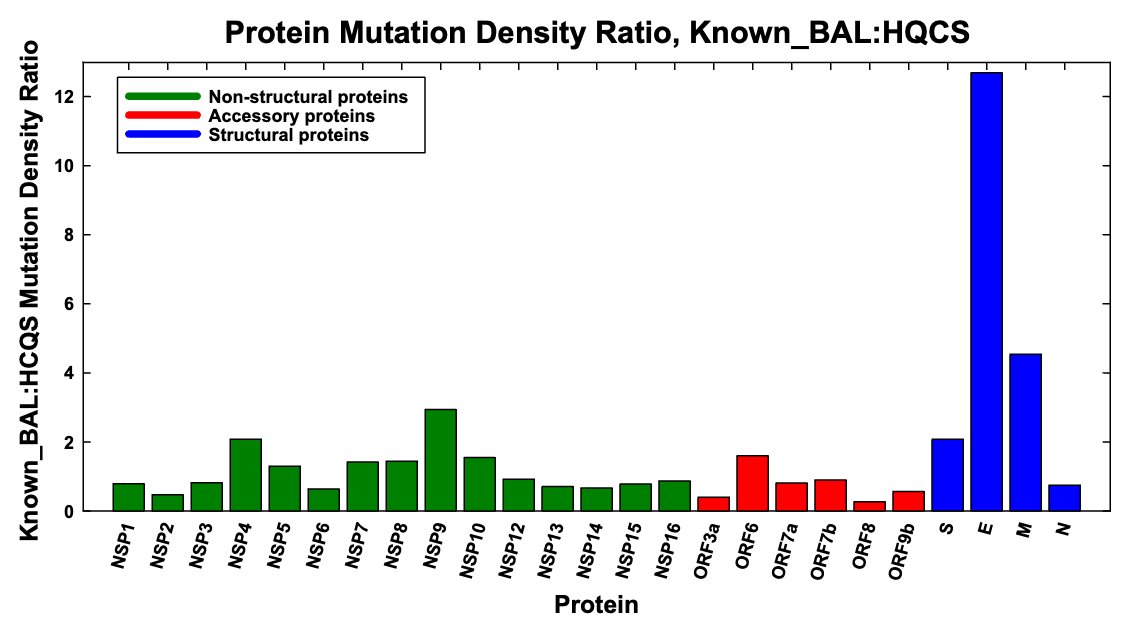
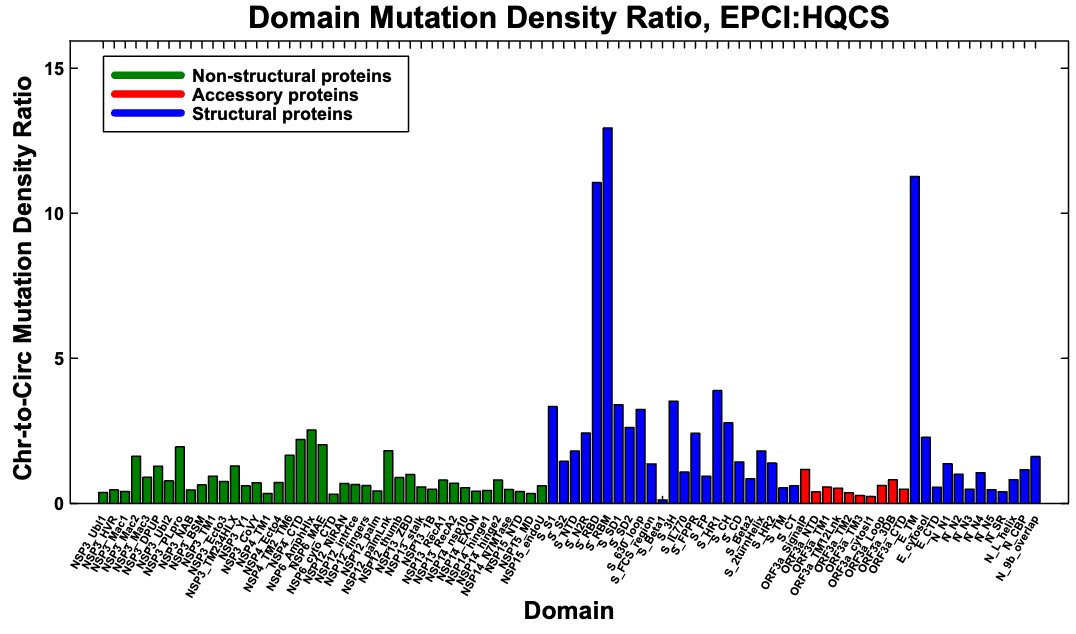
To illustrate how extreme the E mutation rate is in known BAL chronic sequences, below is the ratio of mutation density in each SARS-CoV-2 protein in known, labeled BAL seqs vs high-quality circulating sequences (HCQS).
The wider BAL-pattern graph (~400 seqs) is similar. 12/
The wider BAL-pattern graph (~400 seqs) is similar. 12/
The E mutations are concentrated in the E:5-42 range. I used known protein domains in the graph below, so the outlier is E:8-38. If I'd used E:5-42, it would stand out even more from the rest. I've also included the wider BAL-pattern-sequence graph here. 13/ 
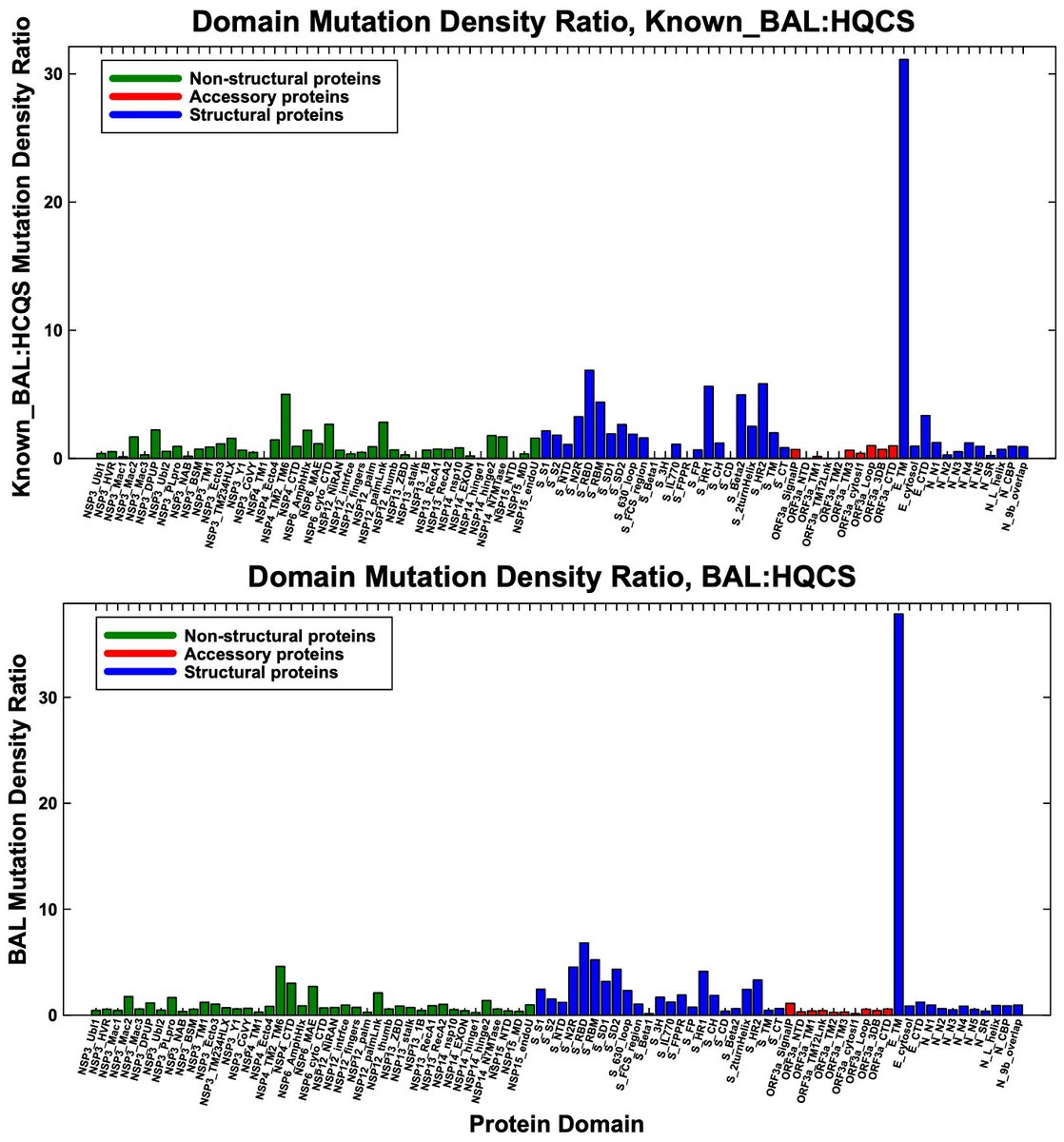
E has the highest mutation-density ratio in the overall chronic-sequence dataset, but this is largely—perhaps mostly—driven by BAL-pattern sequences, and it's not an outlier. Note the stark contrast between RBD/RBM mutation rates in chronics overall vs BAL-only. 14/ 
At first this mutational pattern was similar to the rest—in the sense that I had no inkling of why it occurred. But then I noticed that a number of sequences with this pattern were from bronchoalveolar lavage (BAL) samples, i.e. from deep-lung fluid. 15/
There are 37 independent BAL samples on my list of chronic-infection seqs. All but one has this mutational signature (some more extensively than others). Sequences are often poorly labeled—e.g. the UK includes no info but country & date… 16/
…so I'm certain many sequences with a BAL signature but not labeled BAL are, in fact, BAL samples. One great published case study describes a BAL sequence but the corresponding GISAID sequence gives no indication it was obtained via BAL. 17/
So what does this have to do with Long Covid & the preprint described above? The pattern of antibodies in this Long Covid cohort is exactly what you'd expect to produce the mutational pattern seen in these deep-lung samples: lots of immune pressure on E, very little on spike. 18/
Together with evidence of ongoing activation of mucosal immune responses & the many case studies of symptomatic patients who repeatedly tested negative by nasal swab but test positive on BAL (see 13 below I've run across, likely a fraction of all published)— 19/ 


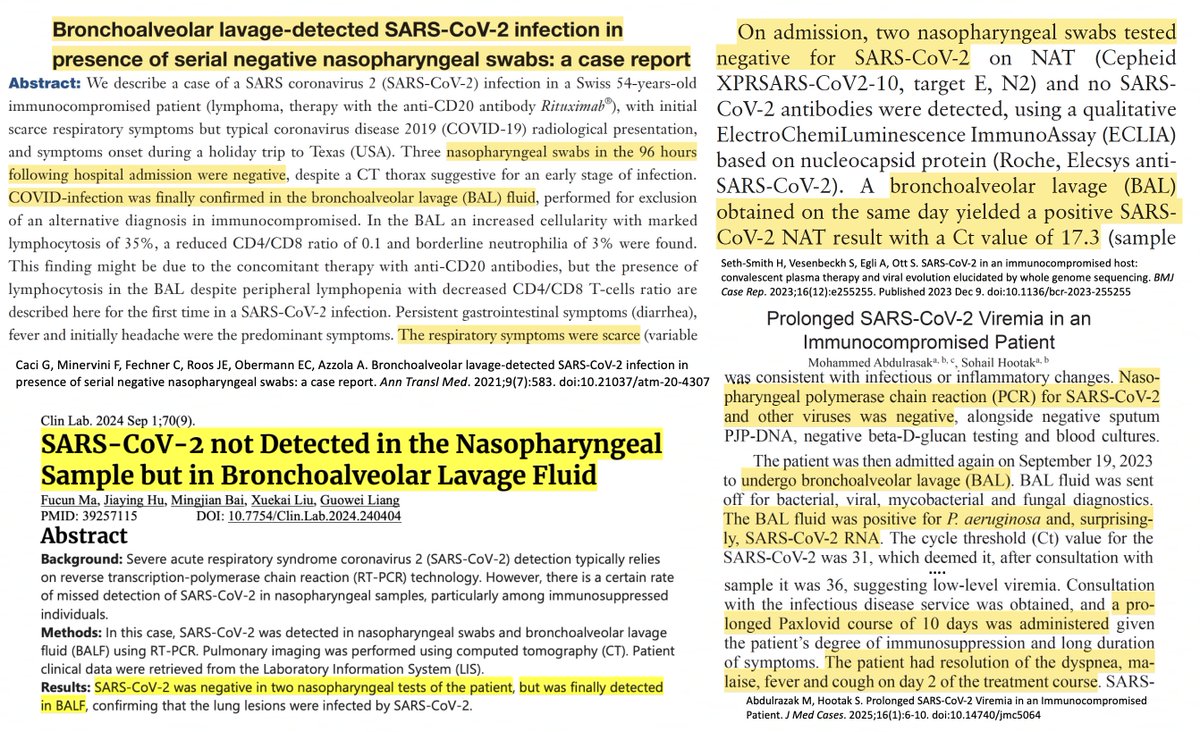


—often cured w/subsequent combination treatment, BTW. I think these results suggest a large proportion of Long Covid cases may be due to chronic infections in the deep lung—or perhaps another tissue resembling the deep lung & producing similar selection pressures. 20/
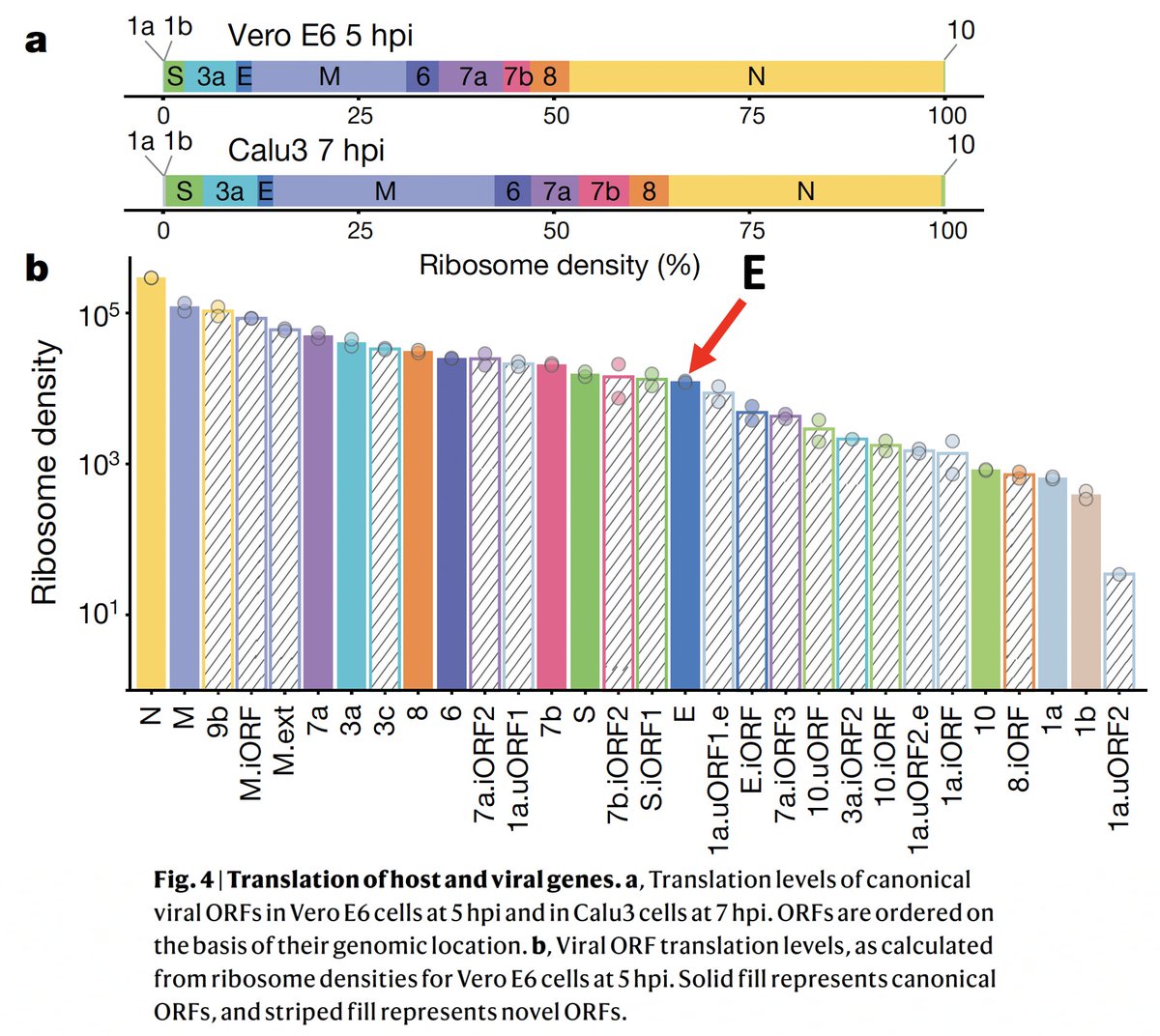
Some may suggest the ongoing immune response is due to viral protein fragments. I've never bought this idea, and in the case of the E protein I think it's especially implausible. E is the least abundantly expressed canonical SARS-CoV-2 protein. 21/
I only know of one study that's measured absolute levels of SARS-CoV-2 protein expression: The Coding Capacity of SARS-CoV-2 in Nature in 2020. They used ribosome profiling, which measures only the mRNA actually being translated into protein at a given moment. 22/ 
E was the least abundantly expressed canonical viral protein—lagging even obscure non-canonical proteins. The notion that the sliver of E produced during acute infection would persist for >6 months, provoking unrelenting immune activation, seems fanciful. 23/ 
A persistent viral reservoir, on the other hand, would continually produce viral proteins, including E, consistent with the near-constant immune activation seen in these LC patients. 24/
It's worth noting that there is at least one case in which there is ironclad genetic evidence of prolonged infection (>1 year), during which the patient had serious respiratory symptoms and lung CT scans characteristic of Covid—& yet tested negative on BAL. 25/ 
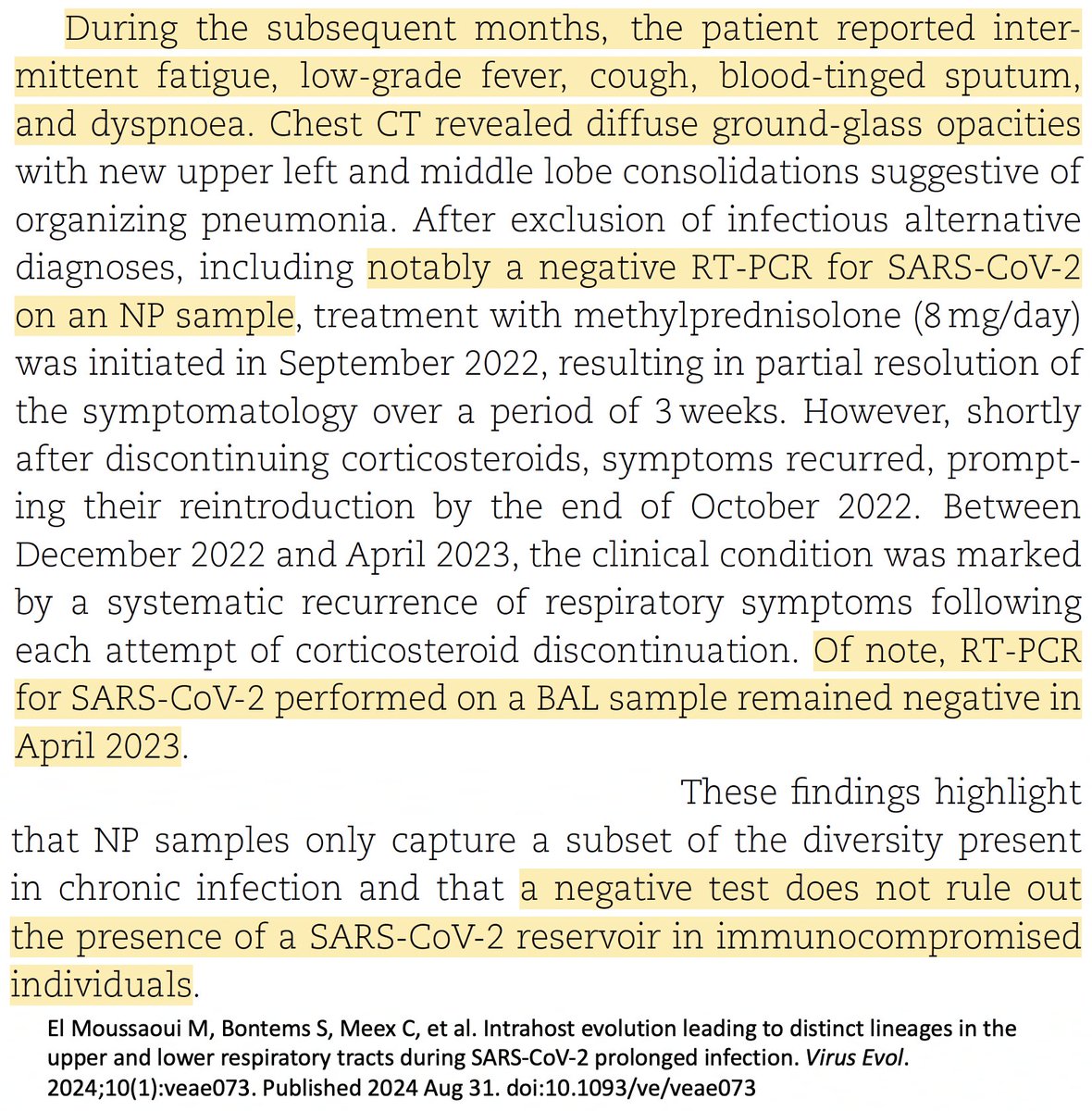
That patient tested positive for Covid a month later, via NP swab & BAL, & recovered after treatment. But after many months of symptoms & repeated negative Covid tests, including via BAL, I imagine many doctors would've stopped testing. 26/academic.oup.com/ve/article/10/…
Deep sequencing of lung (BAL), trachea (ETA), & nose (NP) samples in that patient reveals the expected pattern: many BAL mutations in the BAL sample. Two typical BAL muts were in the NP swab, likely migrating there from an earlier deep-lung intrahost variant. 27/ 

Bronchoalveolar lavage is extremely invasive, so it's only performed upon extreme respiratory distress, which, in the post-acute phase of Covid, is almost always confined to the immunocompromised (IC). 28/
If a persistent, deep-lung viral reservoir is causing the immunological activation & dysfunction of a significant proportion of Long Covid patients, it would likely not be as widespread or uncontrolled (within the lung) as in IC patients, & BAL would not be justified. 29/
In any case, given that even some IC patients w/severe lung pathology clearly caused by Covid have tested negative via BAL (see e.g. above), it's not clear BAL would detect any deep-lung, immune-dysregulating, Long Covid-causing viral reservoir—a frustrating scenario. 30/
Long Covid takes myriad forms & its causes are varied. There's little certainty to be found—except of the suffering it causes & the often unrecognized toll it exacts on society, amply documented by @zalaly, @PutrinoLab, @VirusesImmunity, @EricTopol, @ahandvanish, & others. 31/
@zalaly @PutrinoLab @VirusesImmunity @EricTopol @ahandvanish I don't pretend to know the answers. But there's a large body of evidence suggesting chronic infection can be a cause—see excellent reviews by @microbeminded2 together with @BrodinPetter @johanvawe @EJohnWherry @WesElyMD @MichaelPelusoMD & others 32/ thelancet.com/journals/lanin…
@zalaly @PutrinoLab @VirusesImmunity @EricTopol @ahandvanish @microbeminded2 @BrodinPetter @johanvawe @EJohnWherry @WesElyMD @MichaelPelusoMD This preprint not only adds to the evidence pointing to persistent infection but hints that cryptic viral reservoirs in the deep lung may be involved in LC. Much remains mysterious, but I think this is a possibility worth pursuing. biorxiv.org/content/10.110… 33/end
• • •
Missing some Tweet in this thread? You can try to
force a refresh
















