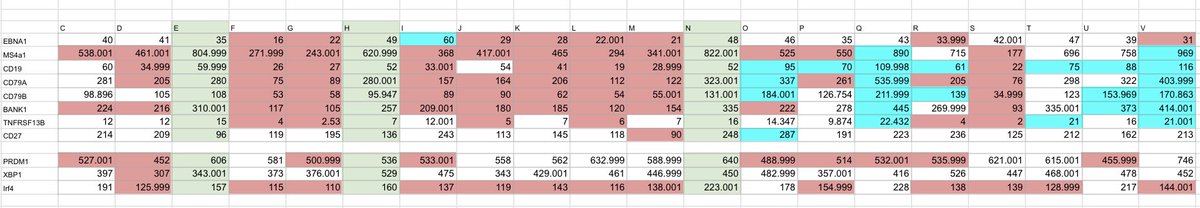
🔬 A large genetic study just found links between specific immune receptor variants (called KIR alleles) and ME/CFS.
And into DecodeME findings.
Simplified breakdown 🧵
And into DecodeME findings.
Simplified breakdown 🧵

ME/CFS is a disabling disease that affects energy, memory, sleep, and physical function.
Many cases begin after an infection.
Its biological causes are not completely known, but immune system problems - especially involving NK cells - are a main theory.
Many cases begin after an infection.
Its biological causes are not completely known, but immune system problems - especially involving NK cells - are a main theory.
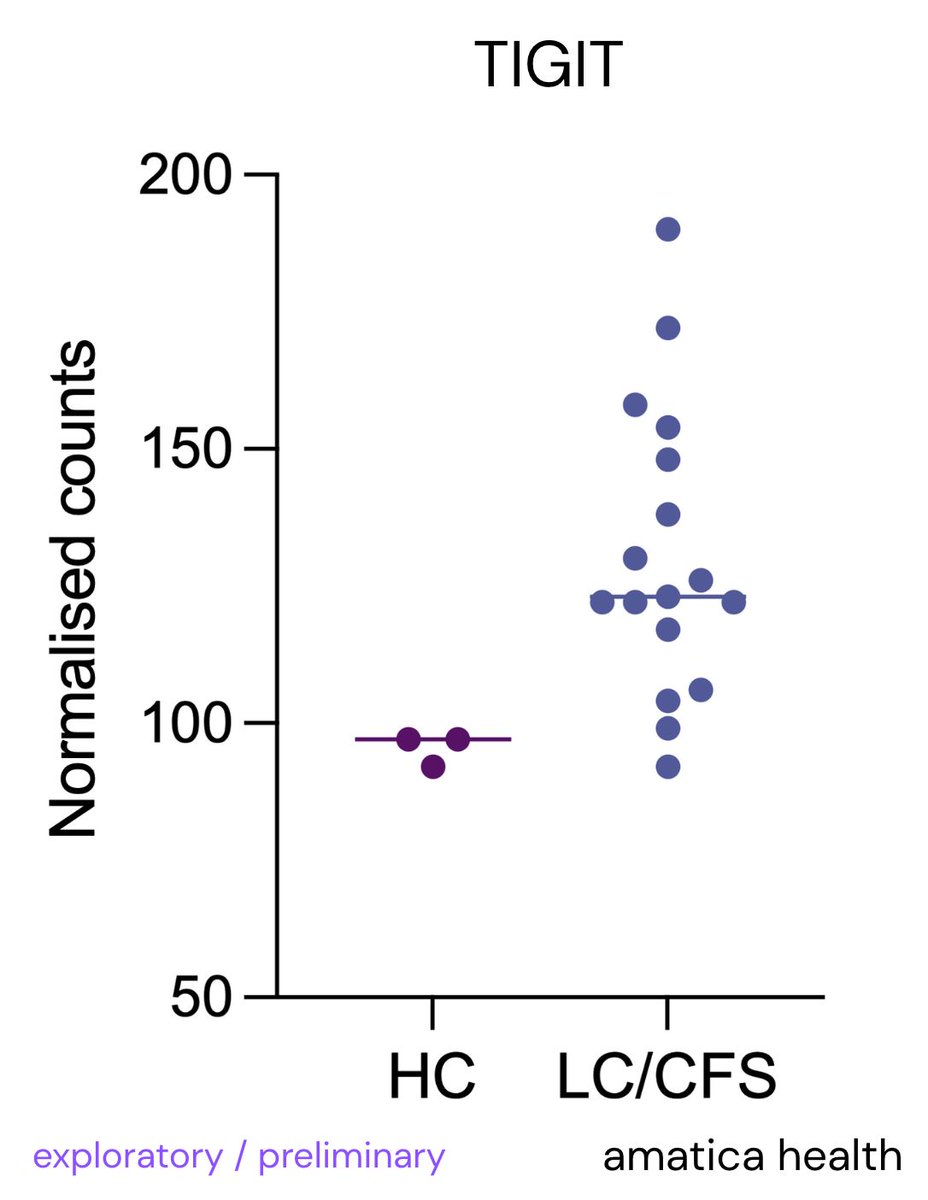
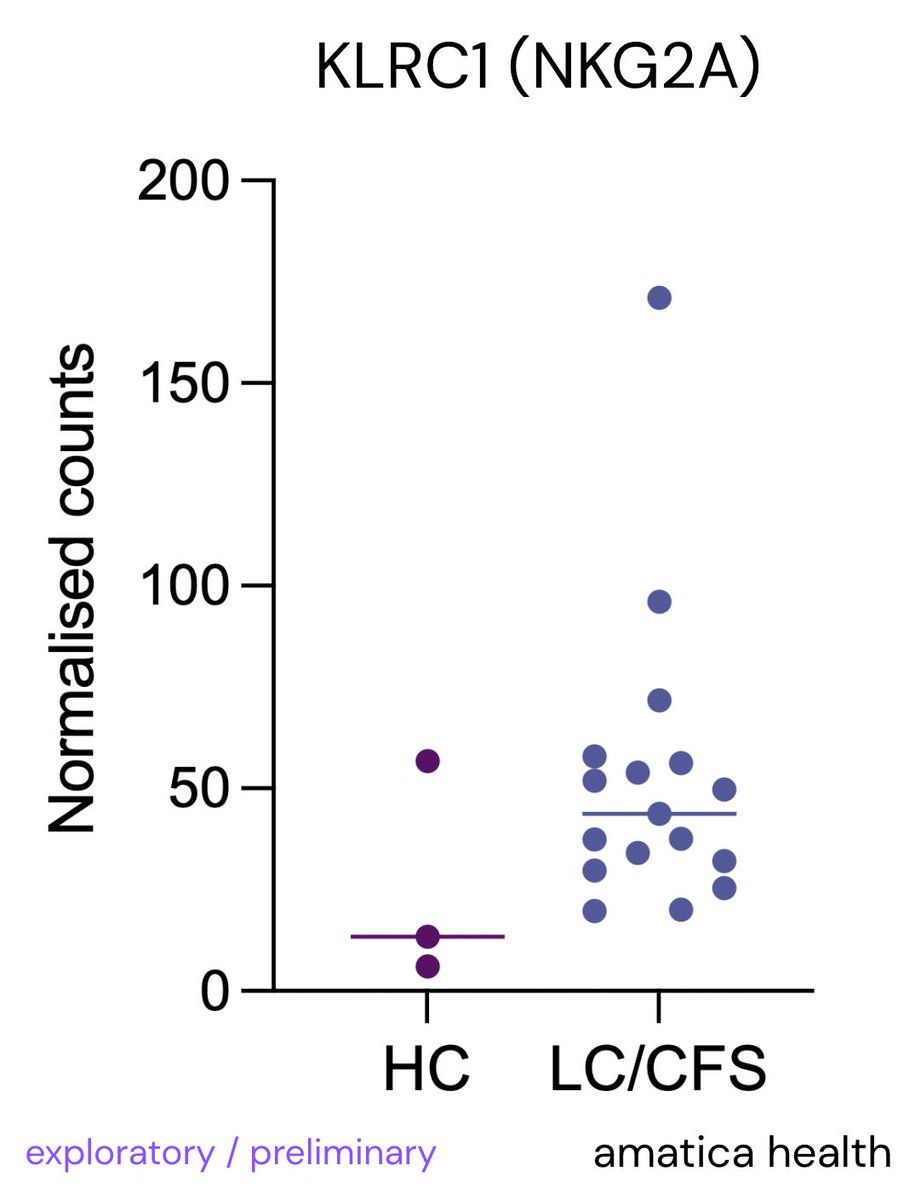
We @amaticahealth have also found raised NKG2A & TIGIT (suppressive markers on NK cells & hinder immune response)
You can get info on your NK cell function via testing:
amaticahealth.com/me-cfs-long-co…

You can get info on your NK cell function via testing:
amaticahealth.com/me-cfs-long-co…
@amaticahealth Natural killer (NK) cells are part of the innate immune system.
They help detect and eliminate infected or abnormal cells.
Their activity is regulated by receptors on their surface - one important family is called KIR (Killer-cell Immunoglobulin-like Receptors).
They help detect and eliminate infected or abnormal cells.
Their activity is regulated by receptors on their surface - one important family is called KIR (Killer-cell Immunoglobulin-like Receptors).
@amaticahealth KIRs come in many genetic variants.
Some are "inhibitory" (they tell NK cells to stop), others are "activating" (they say go).
These receptors work by recognizing markers (HLA molecules) on other cells.
Everyone has a different set of KIRs and HLAs.
Some are "inhibitory" (they tell NK cells to stop), others are "activating" (they say go).
These receptors work by recognizing markers (HLA molecules) on other cells.
Everyone has a different set of KIRs and HLAs.
@amaticahealth Researchers studied 418 Norwegian ME/CFS patients and 473 healthy people, analyzing their KIR genes in very high detail.
This is the largest and most detailed KIR study ever done in ME/CFS.
This is the largest and most detailed KIR study ever done in ME/CFS.
@amaticahealth First key finding:
Just having or not having certain KIR genes wasn’t different between ME/CFS patients and controls.
No differences in KIR gene “content” or “copy number” were found.
Just having or not having certain KIR genes wasn’t different between ME/CFS patients and controls.
No differences in KIR gene “content” or “copy number” were found.
@amaticahealth But when the researchers looked at specific versions of KIR genes - called alleles - they found clear differences between patients and controls.
@amaticahealth Three KIR alleles were more frequent in ME/CFS patients:
- KIR3DL3002
- KIR3DL1020
- KIR3DL2*009
Each of these was associated with about a 1.4–2.2x higher risk of ME/CFS.
- KIR3DL3002
- KIR3DL1020
- KIR3DL2*009
Each of these was associated with about a 1.4–2.2x higher risk of ME/CFS.
@amaticahealth Two KIR alleles were less frequent in ME/CFS patients:
- KIR3DL3013
- KIR3DL2010
These may have a protective effect, reducing the odds of ME/CFS by about half.
- KIR3DL3013
- KIR3DL2010
These may have a protective effect, reducing the odds of ME/CFS by about half.
@amaticahealth All five of these genes encode inhibitory receptors.
These are receptors that tell immune cells not to attack.
This suggests ME/CFS patients might have stronger-than-usual “braking signals” in their immune system.
These are receptors that tell immune cells not to attack.
This suggests ME/CFS patients might have stronger-than-usual “braking signals” in their immune system.
@amaticahealth In fact, KIR3DL1*020 - one of the alleles more common in ME/CFS - is known to be a high-expression variant.
That means it makes more protein and gives stronger inhibition signals to NK cells.
Similar to our preliminary findings with increased NKG2A & TIGIT.
That means it makes more protein and gives stronger inhibition signals to NK cells.
Similar to our preliminary findings with increased NKG2A & TIGIT.
@amaticahealth KIR3DL2009, another risk allele, is thought to have lower expression.
That could mean weaker ability to respond to certain pathogen signals.
So while KIR3DL1020 may over-inhibit, KIR3DL2*009 might under-sense infections.
That could mean weaker ability to respond to certain pathogen signals.
So while KIR3DL1020 may over-inhibit, KIR3DL2*009 might under-sense infections.
@amaticahealth Some of these risk and protective alleles appear together on specific haplotypes - groups of gene variants inherited together.
A few of these haplotypes were significantly more or less common in ME/CFS patients.
A few of these haplotypes were significantly more or less common in ME/CFS patients.
@amaticahealth The strongest was a haplotype containing KIR3DL1020 and KIR3DL2009.
It was twice as common in ME/CFS patients compared to controls.
That combination may tip the immune balance.
It was twice as common in ME/CFS patients compared to controls.
That combination may tip the immune balance.
@amaticahealth Researchers also looked at the HLA ligands that interact with KIRs.
One particular ligand - called Bw4-80I, part of certain HLA-B types - was more common in patients.
One particular ligand - called Bw4-80I, part of certain HLA-B types - was more common in patients.
@amaticahealth Importantly, people who had both KIR3DL1 and Bw4-80I were more likely to have ME/CFS.
This is a known receptor-ligand pair, and their combined presence may over-suppress NK cell function.
This is a known receptor-ligand pair, and their combined presence may over-suppress NK cell function.
@amaticahealth This KIR3DL1-Bw4-80I combination has been seen in other diseases too - like psoriasis and autoimmune hepatitis - but the impact seems to vary depending on the exact alleles involved.
@amaticahealth Another interesting point:
KIR3DL2 isn’t just on NK cells.
It’s also found on some T cells and can detect viral or bacterial DNA (CpG sequences).
Risk allele KIR3DL2*009 may be less effective at that.
KIR3DL2 isn’t just on NK cells.
It’s also found on some T cells and can detect viral or bacterial DNA (CpG sequences).
Risk allele KIR3DL2*009 may be less effective at that.
@amaticahealth KIR3DL3 - one of the associated genes - has only recently been linked to immune function.
It’s found on T cells like γδ T cells and interacts with a molecule called HHLA2 (not HLA).
Its role in ME/CFS is unclear but intriguing.
It’s found on T cells like γδ T cells and interacts with a molecule called HHLA2 (not HLA).
Its role in ME/CFS is unclear but intriguing.
@amaticahealth All this adds to the idea that ME/CFS involves immune dysregulation, possibly in response to infections. Which supports leading theories.
Around 73% of ME/CFS patients in this study said their illness began after an infection.
Around 73% of ME/CFS patients in this study said their illness began after an infection.
@amaticahealth The KIR genes may influence how NK and T cells react to infections - either failing to clear the virus or overreacting in a way that causes chronic symptoms.
Both scenarios could help explain ME/CFS onset.
Both scenarios could help explain ME/CFS onset.
@amaticahealth These results don’t prove causation and need to be replicated in larger studies.
But they offer a concrete genetic link between immune receptor variation and ME/CFS - and a plausible path forward for deeper investigation.
But they offer a concrete genetic link between immune receptor variation and ME/CFS - and a plausible path forward for deeper investigation.
• • •
Missing some Tweet in this thread? You can try to
force a refresh