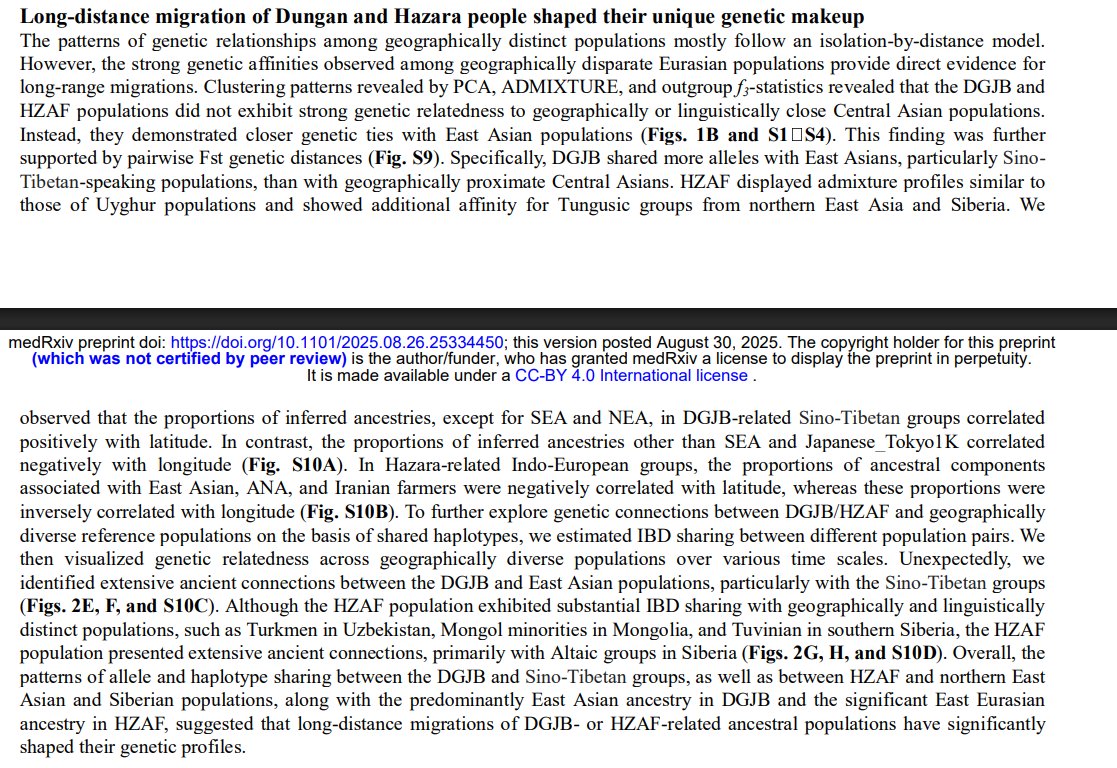
Whole-Genome Sequencing Pilot Study of the Central Asian Genetic Diversity Project Reveals Distinct Genetic Histories, Adaptive Processes, and Introgression Events
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…

The phenotypic effects of the identified high-confidence Neanderthal- and Denisovan-derived segments were systematically examined. 
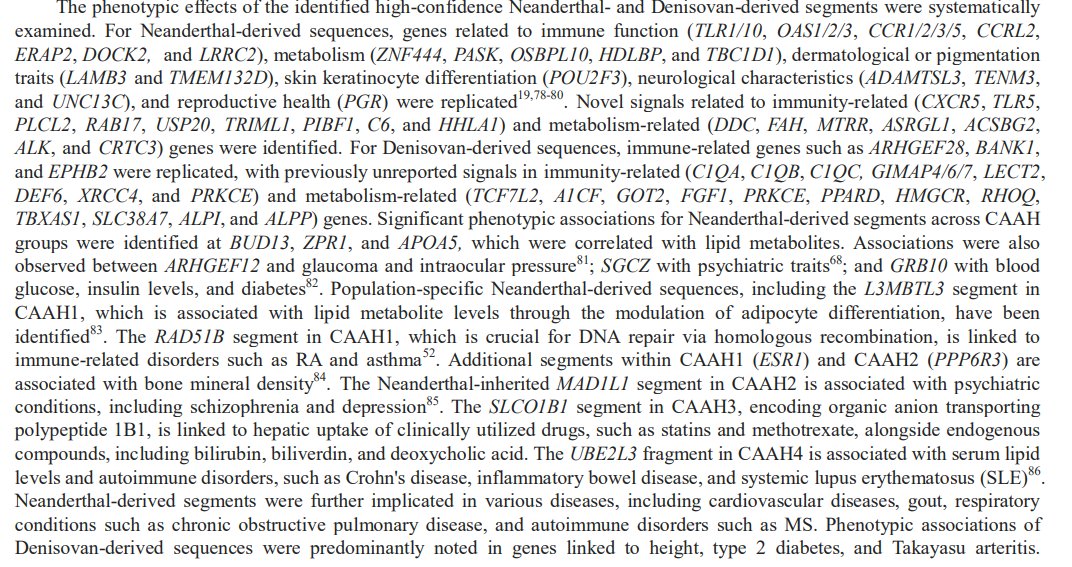
Additionally, archaic variants in HHAT (rs115453328) identified in CAAH1 were associated with obsessive-compulsive traits 
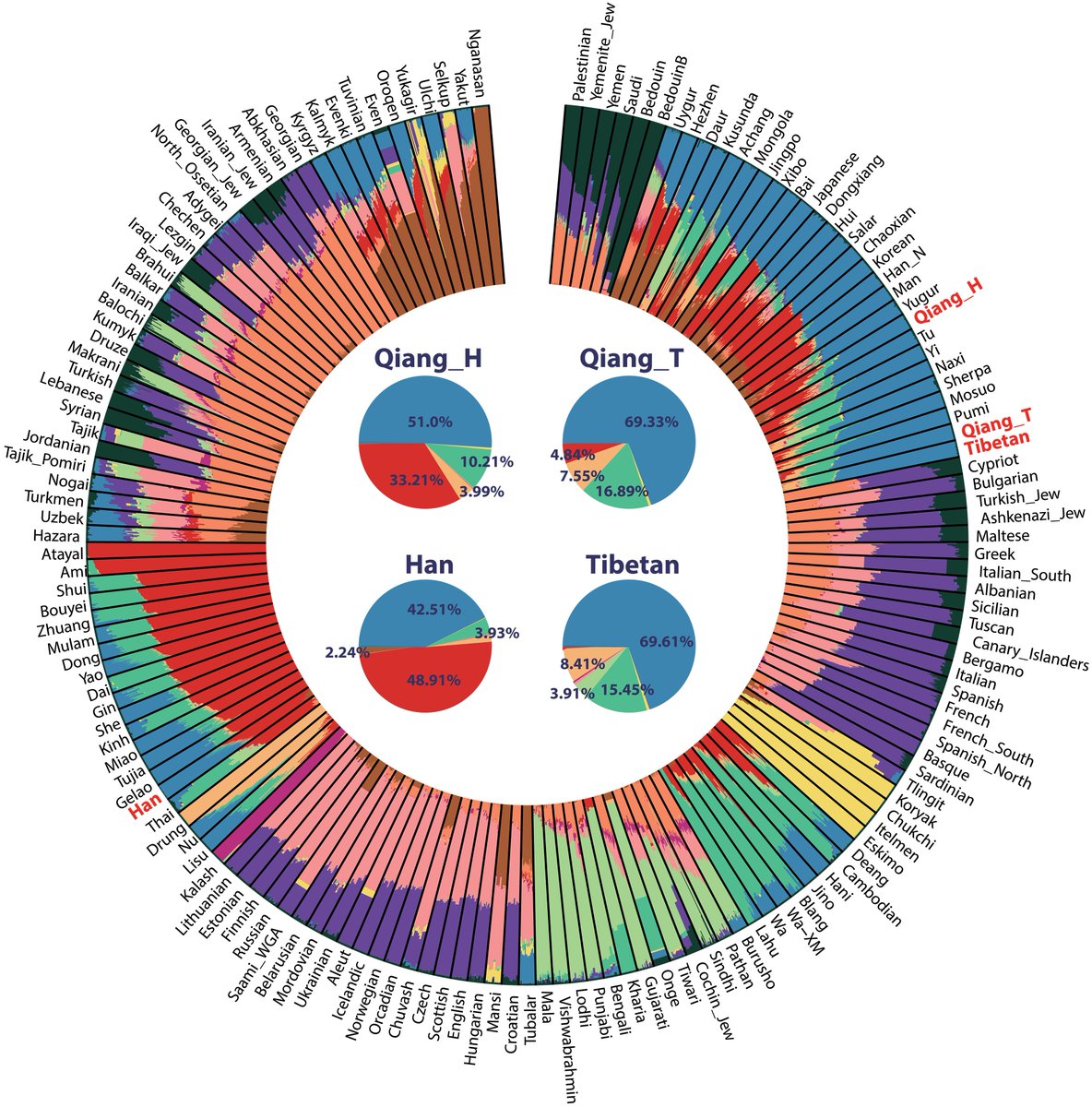
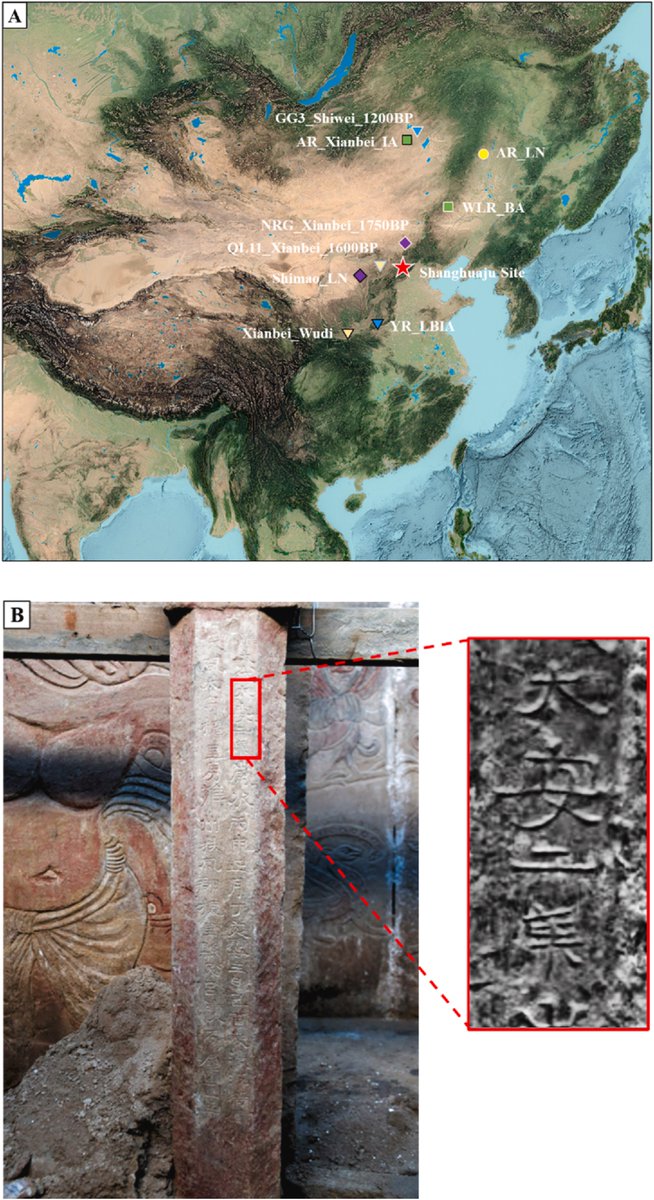
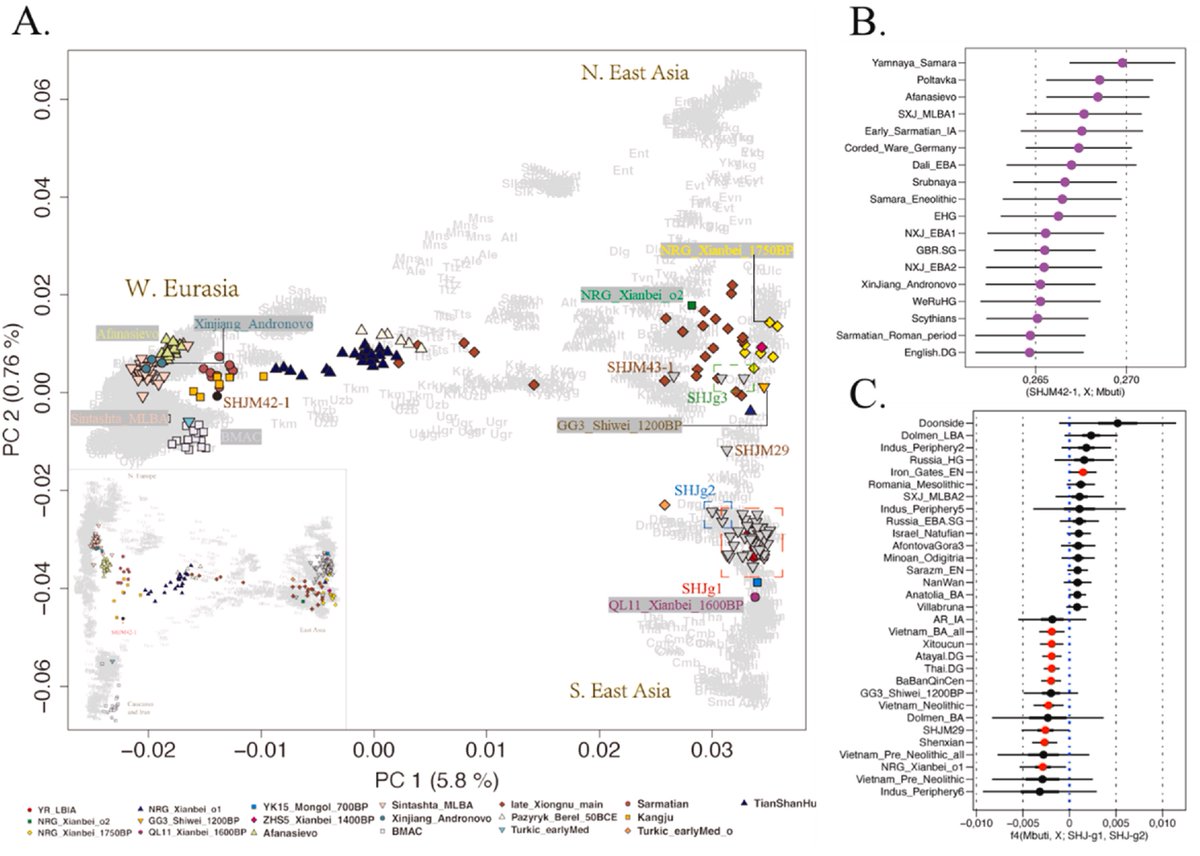
The differentiated genetic architecture of linguistically close Turkic, Indo-European, and Sino-Tibetan groups 

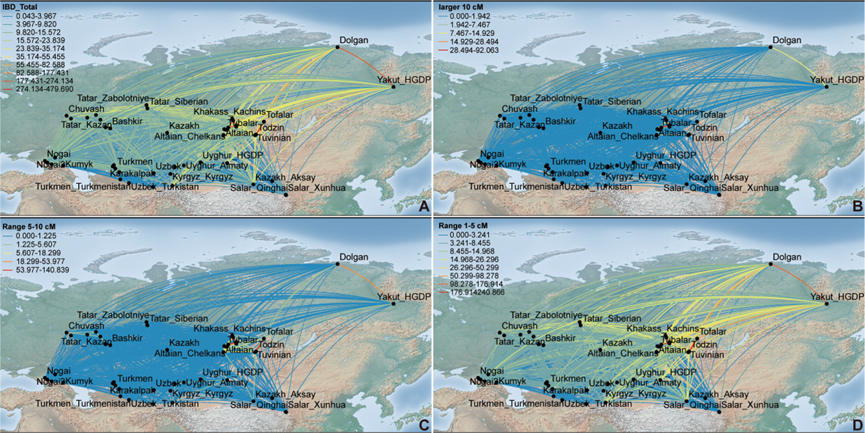
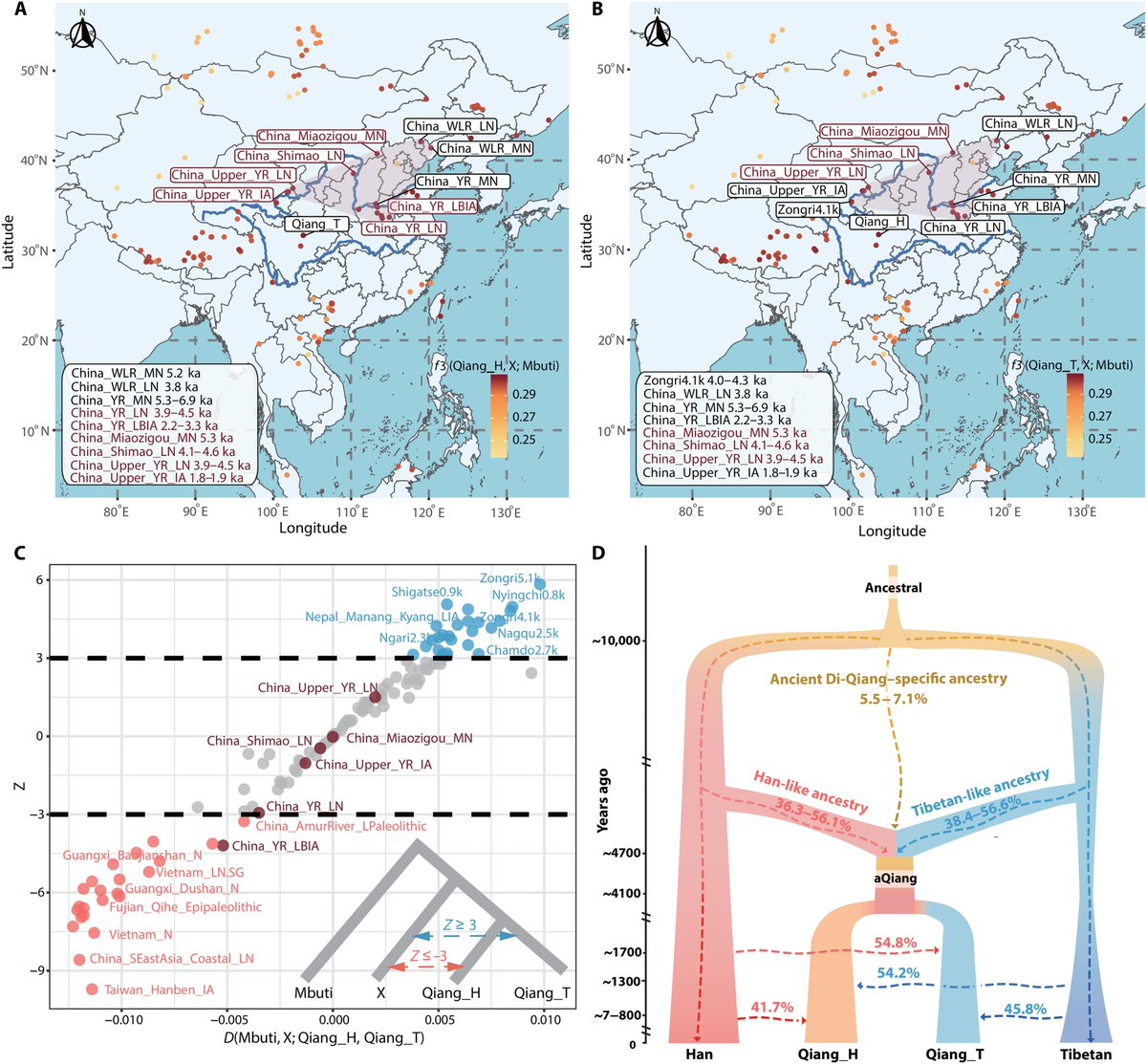
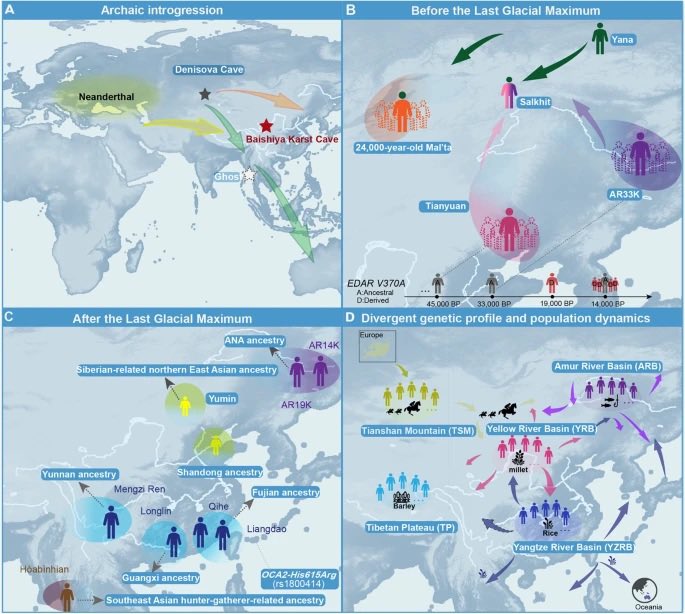
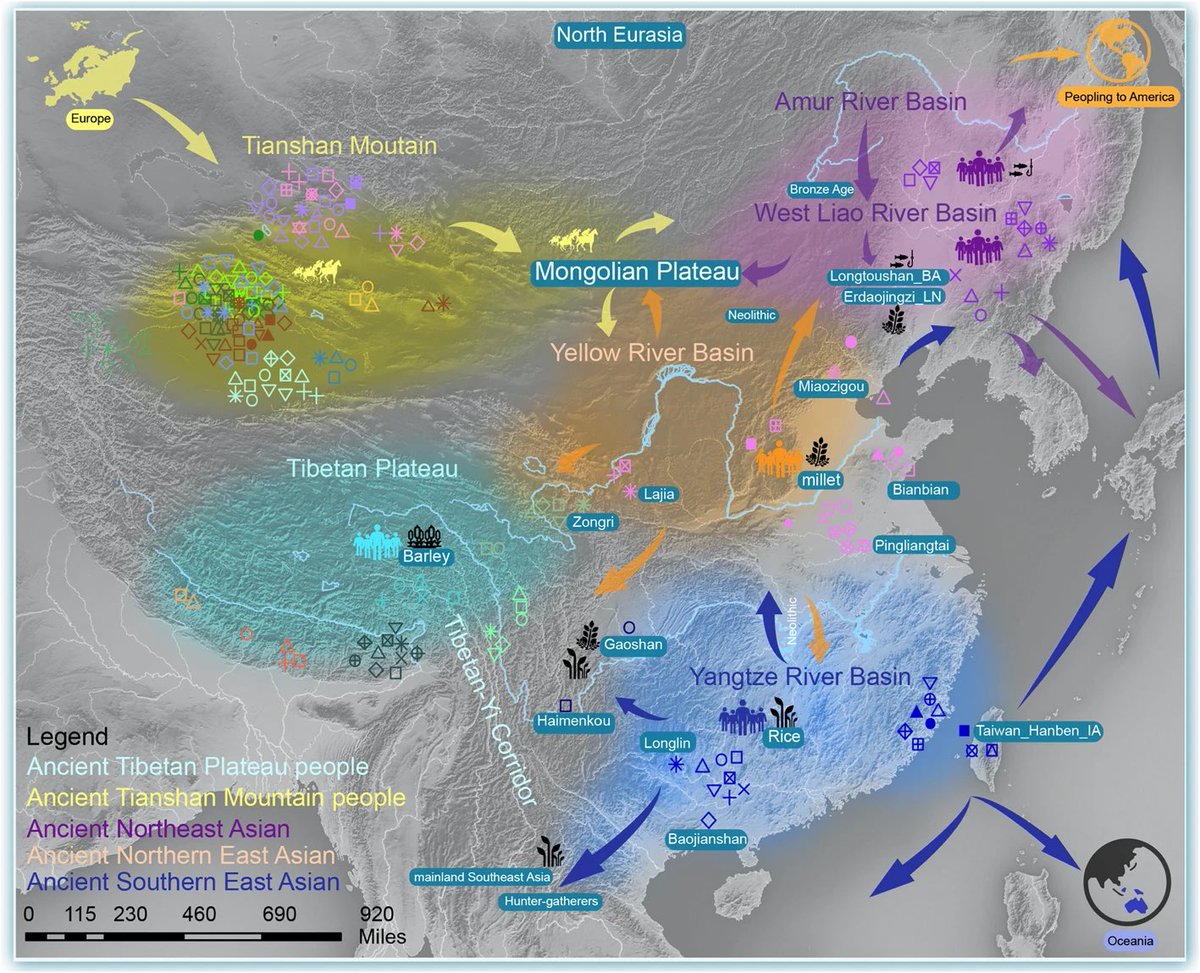
Genetic relationships between newly sequenced CAAH and spatiotemporally diverse Eurasian reference populations revealed based on the merged Human Origins (HO) dataset. (A) 

• • •
Missing some Tweet in this thread? You can try to
force a refresh