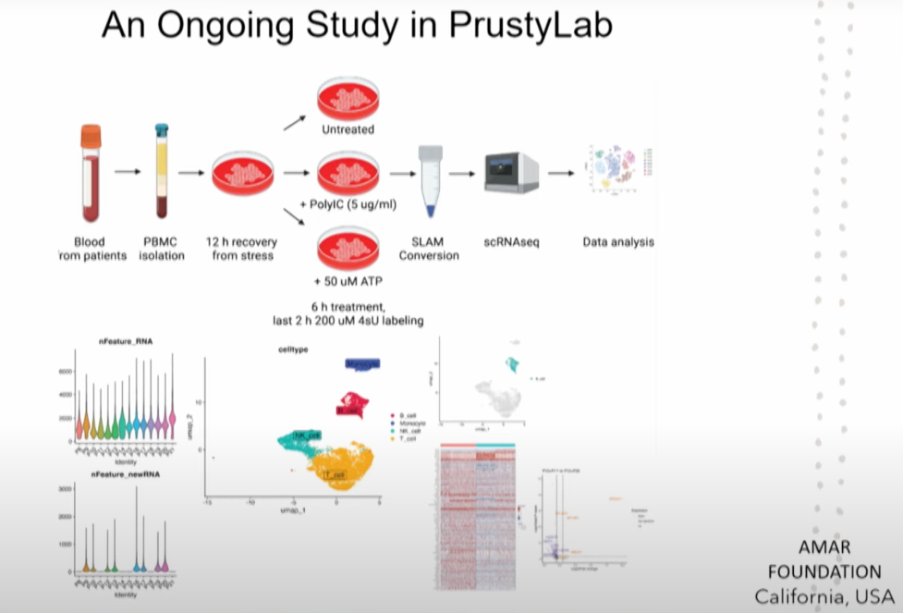
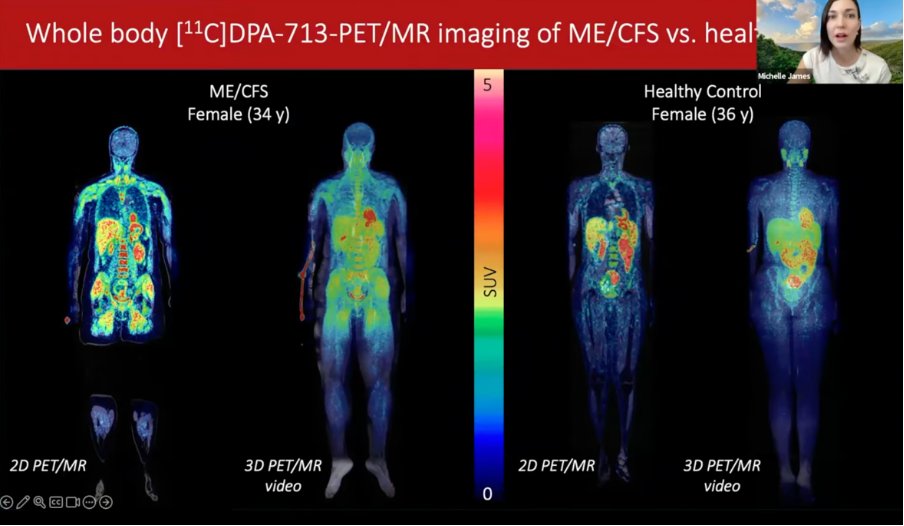
1) The most interesting presentation during the 2025 Stanford symposium was on PET scans of the entire body.
Dr. Michelle James explained that they found a striking pattern with more TSPO signal in various muscle groups of ME/CFS patients, such as the thigh and shoulders.
Dr. Michelle James explained that they found a striking pattern with more TSPO signal in various muscle groups of ME/CFS patients, such as the thigh and shoulders.
2) PET scans work by injecting a radioactive molecule into the veins. The scan records annihilation events where a positron leaves the cell and collides with an electron, creating two photons. That signal tells us where in the body the radioactive molecule is binding. 
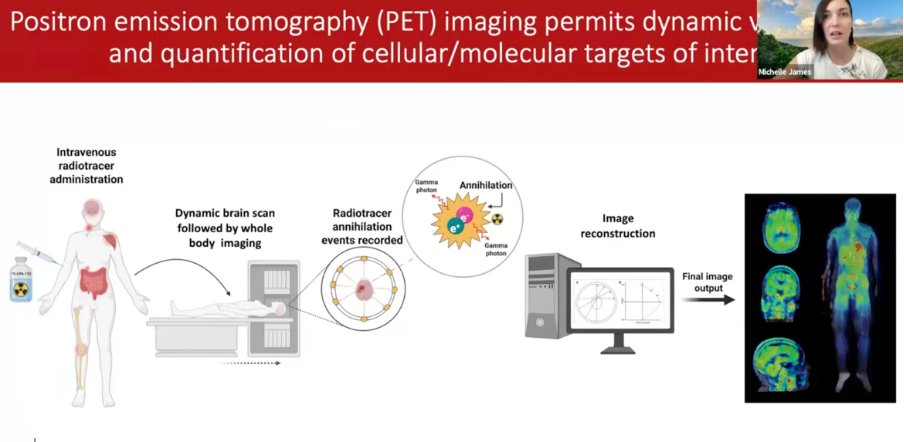
3) Ideally, you want a radioactive tracer that is highly specific to a certain cell type or process so that you know exactly what the signal means.
In this ME/CFS study, they used the popular TSPO tracer, which binds to microglia, astrocytes, myeloid cells, etc.
In this ME/CFS study, they used the popular TSPO tracer, which binds to microglia, astrocytes, myeloid cells, etc.
4) In particular, they used the molecule [11C]DPA-713. It’s used as a marker of inflammation but also signals mitochondrial function and cellular bioenergetics. It has a half-life of only 20 minutes (meaning the signal is gone after around 3 hours).
5) Participants in the study had a 60-minute brain PET scan, followed by a 20-30 min whole body scan.
ME/CFS patients had significantly more signal in muscle groups such as the upper and lower thigh and the glute minimus (see results in screenshot below).
ME/CFS patients had significantly more signal in muscle groups such as the upper and lower thigh and the glute minimus (see results in screenshot below).
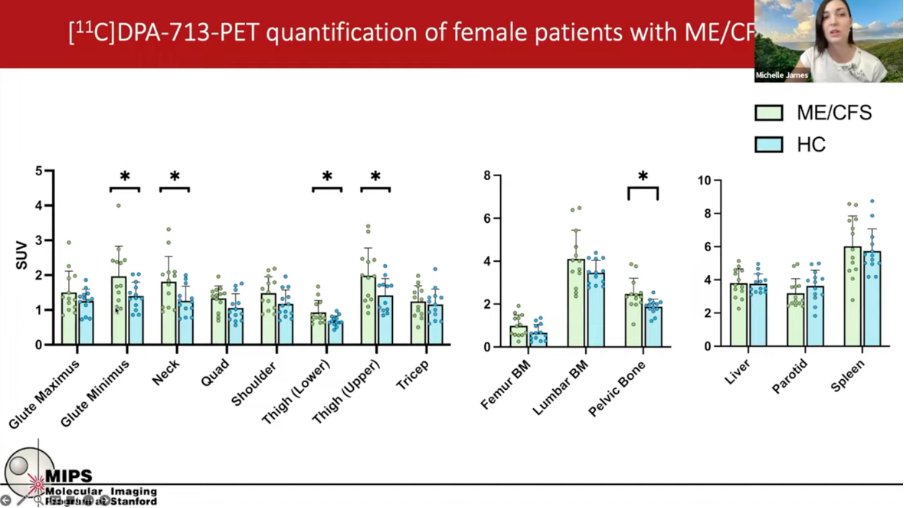
6) There was also a notable pattern around the shoulders which looked like coathanger syndrome. The signals correlated with measures of fatigue (MFI) or pain (BPI).
7) Dr. James said they are working on more specific tracers that target GPR84 and TREM1 on pro-inflammatory myeloid cells. These could be used in ME/CFS studies in the near future and may provide further insights.
8 ) The full presentation by Dr. Michelle James about the PET-scan study can be viewed here:
• • •
Missing some Tweet in this thread? You can try to
force a refresh