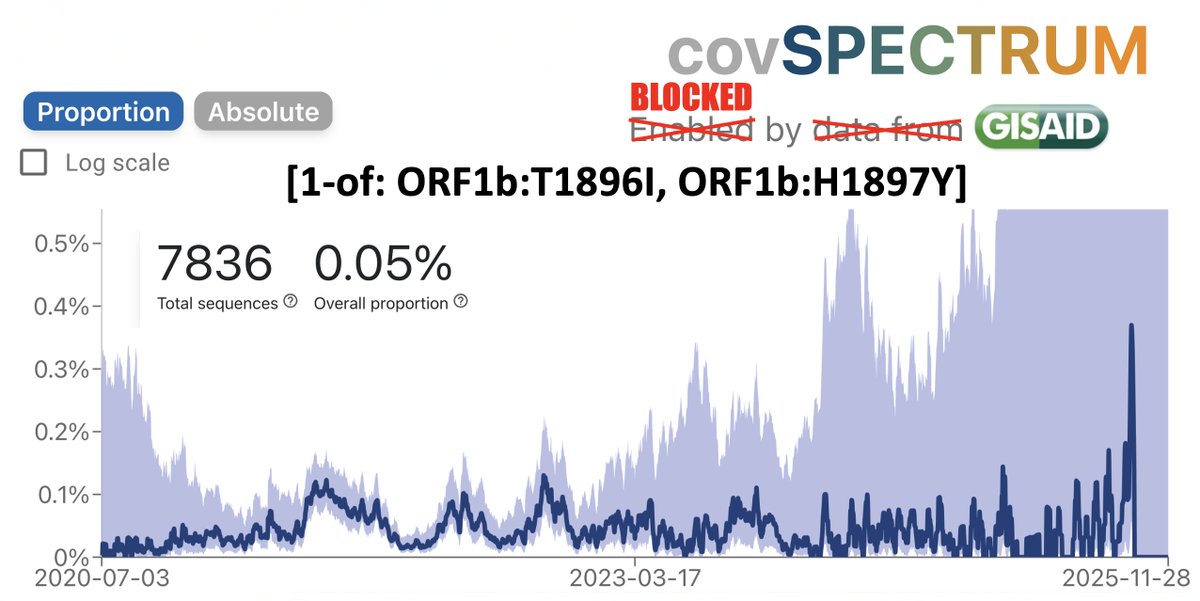
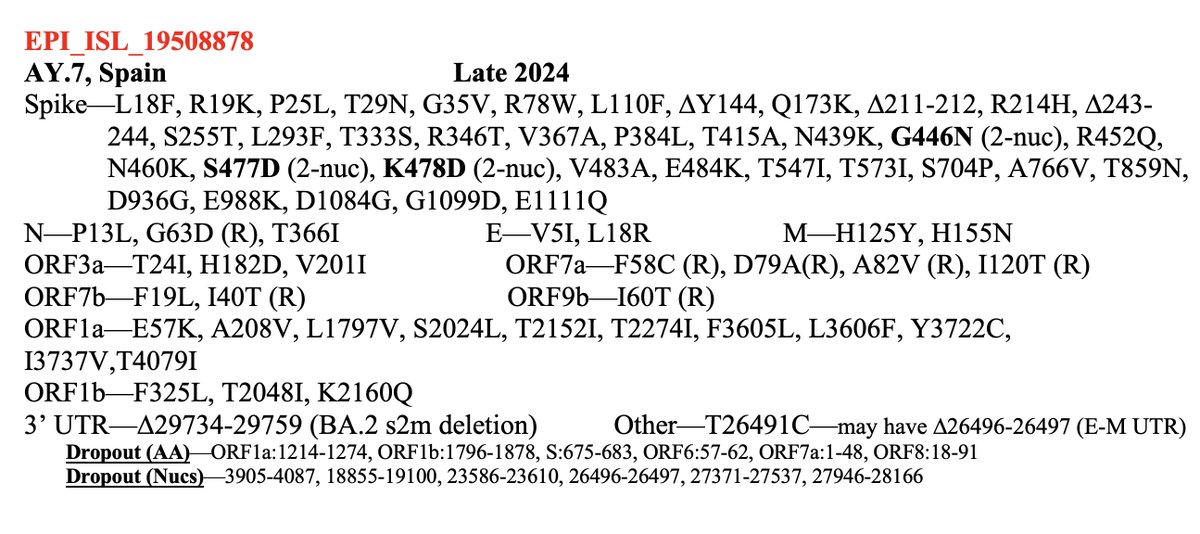
I beg to differ! If it is not a sequencing mistake—and it looks clean—one of these BA.3.2 has something completely novel in SARS-CoV-2 evolution: an FCS-adjacent deletion!
One of the two QT repeats appears to have been deleted. I've never seen anything like this before.
One of the two QT repeats appears to have been deleted. I've never seen anything like this before.
https://twitter.com/snpoehlm/status/1980953067777368569
Work by @TheMenacheryLab looked at a similar, more extensive, deletion. They deleted both QT repeats plus the next AA (∆QTQTN). In Vero cells (monkey kidney cells), it produced extra-large plaques & outcompeted WT virus—similar to furin cleavage site (FCS)-deletion mutants. 2/12 
But in human lung cancer (Calu3) cells, the ∆QTQTN-mutant replication was dramatically reduced (2.5 orders of magnitude), and in infected hamsters disease was much milder. 3/12 
Oddly, despite reduced illness and demonstrably reduced cell entry in vitro, viral replication was apparently increased in hamsters in the nose and throat (as measured by viral RNA titers), while lung replication was unchanged.
Hard to make sense of. 4/12
Hard to make sense of. 4/12

What's certain is that the ∆QTQTN deletion reduces (furin) S1/S2 cleavage. The smaller ∆QT in this BA.3.2 (assuming it is real, as it appears to be), likely has a similar effect.
Would love to hear @StuartTurville's take on this.
5/12
Would love to hear @StuartTurville's take on this.
5/12

Importantly, there's never been a variant with a deletion like this. Though there were a few early reports of FCS-adjacent deletions, I'm skeptical they were real. All were from the same lab, led by a fraudulent scientist (Didier Raoult) who's since had 48 papers retracted. 6/12 

Also, while the FCS lab experiments are immensely valuable, they took place on an ancestral spike background. BA.3.2 has (relative to WT) N679R & P681H. P681H has been shown many times over to increase spike cleavage, and N679R almost certainly also has that effect. 7/12 

Furthermore, BA.3.2 has huge deletions in the spike NTD (S:1-306). The resulting shorter NTD loops (also in SARS-1) have been shown by @EnyaQing to dramatically increase spike cleavage and infectivity (and instability). 8/12
Old 🧵 on this topic:
Old 🧵 on this topic:
https://x.com/LongDesertTrain/status/1569025027701612544
Regions around the FCS have been completely reconfigured in BA.3.2. On top of Omicron's H655Y (which increases time spent in intermediate fusion configurations), it has K529N, E554D, A575S, E583D, H625R, N641K, V642G, and E654K upstream & A688D + S704L downstream of the FCS. 9/12 

And further upstream, in S2, BA.3.2 has two extraordinarily rare & meaningful mutations—S:K795T & the 2-nuc S:A852K.
So the effect of this new FCS-adjacent deletion on infectivity & tropism may be completely different than on a WT spike background. 10/12
So the effect of this new FCS-adjacent deletion on infectivity & tropism may be completely different than on a WT spike background. 10/12
https://x.com/LongDesertTrain/status/1899647074951368860
Finally, this deletion, which likely decreases S1/S2 cleavage, may be part of a larger trend in SARS-CoV-2 evolution. Recently there seems to be real selection pressure for mutations that temper S1/S2 cleavage. It's not clear why (incr stability?) 11/12
https://x.com/LongDesertTrain/status/1971365537906381090
In any case, while the precise consequences of all this aren't known, one thing is clear: BA.3.2 is very different from past variants, none of which could tolerate a deletion like this. What it means going forward is anyone's guess. 12/12
• • •
Missing some Tweet in this thread? You can try to
force a refresh