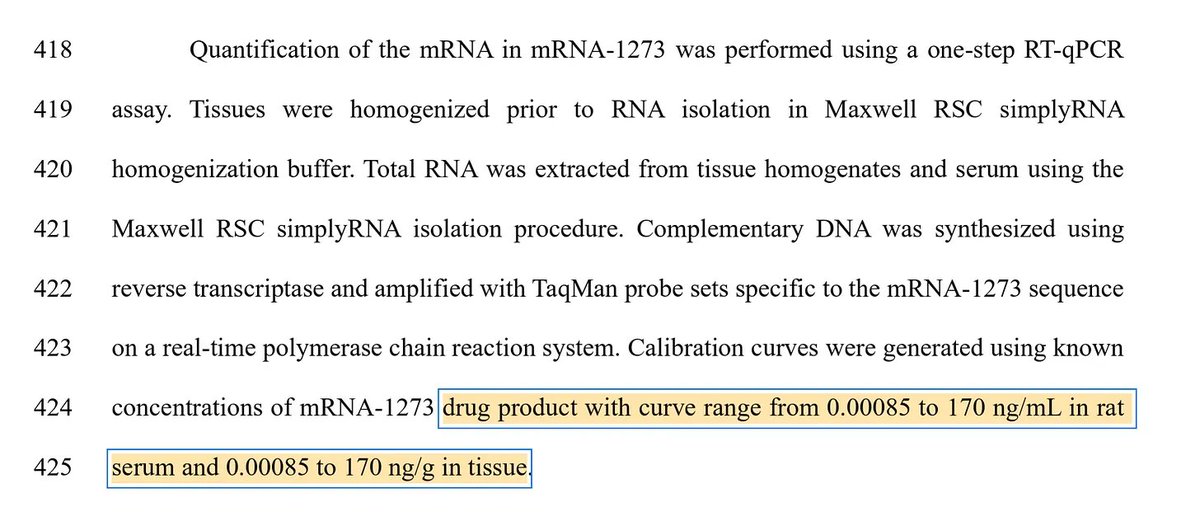
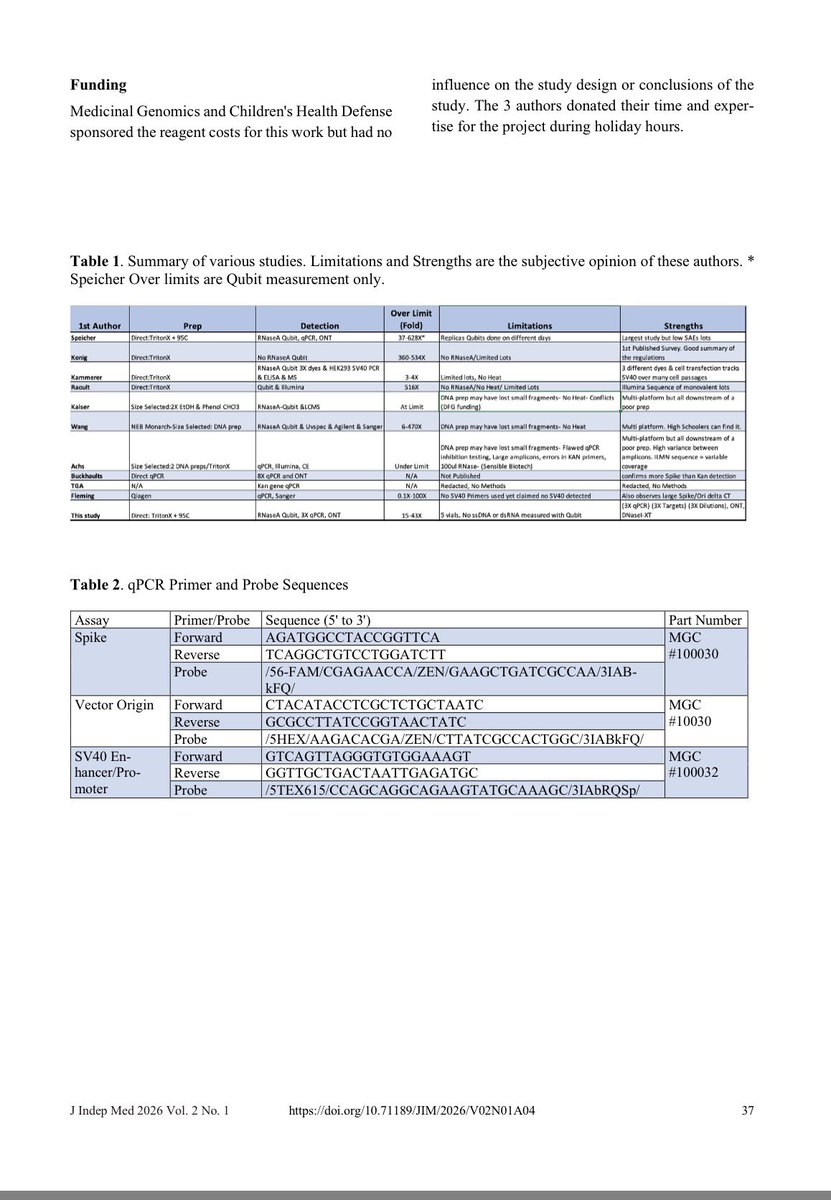
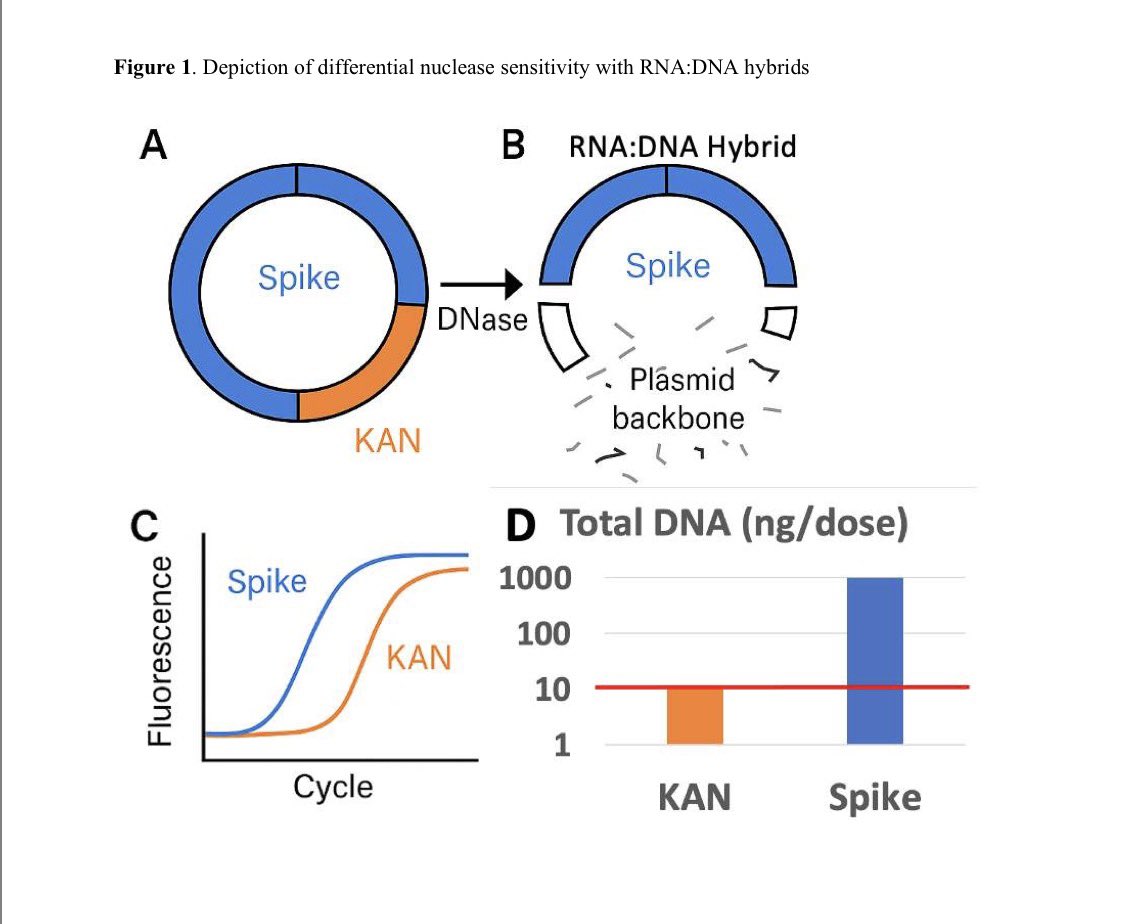
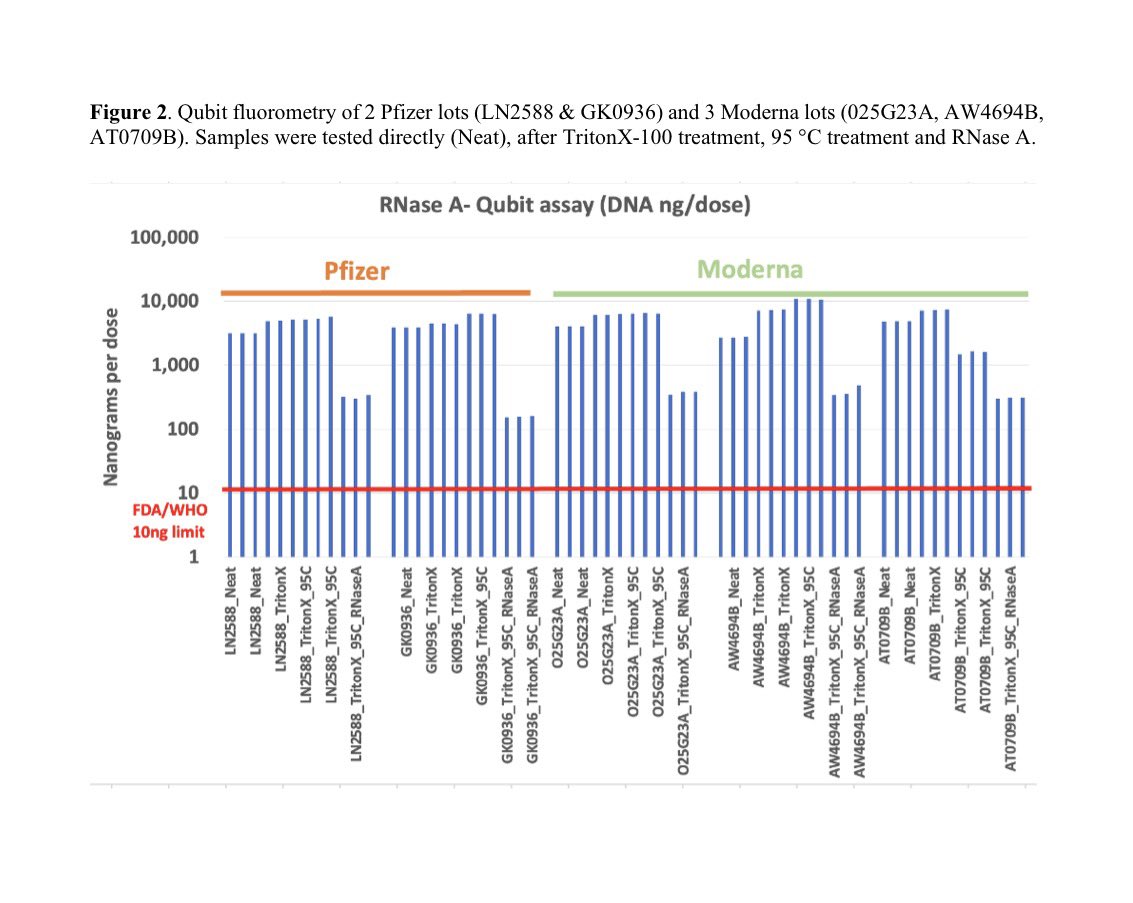
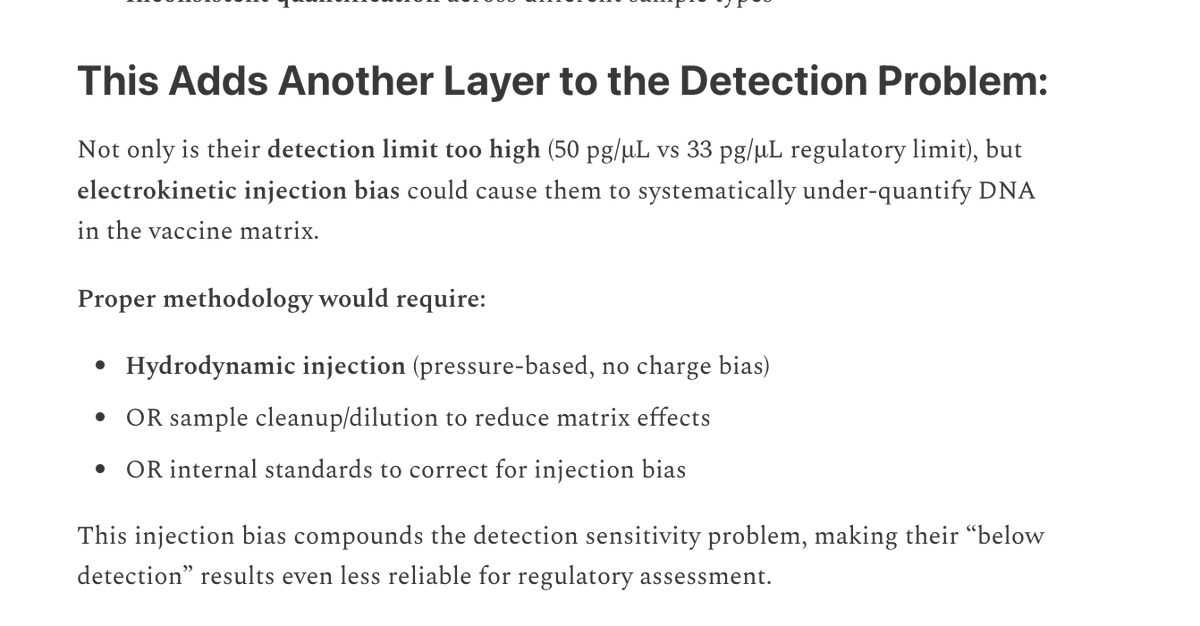
This paper tries to makes claims of RT-qPCR sensitivity with a LLOQ of 372,800 molecules/ml as their lowest limit of Quantiation.
As a comparison, most clinical HIV RT-qPCR tests pick up 50 molecules/ml.
Hence they have very optimistic clearance rates.
biorxiv.org/content/10.648…
As a comparison, most clinical HIV RT-qPCR tests pick up 50 molecules/ml.
Hence they have very optimistic clearance rates.
biorxiv.org/content/10.648…
Check for yourself
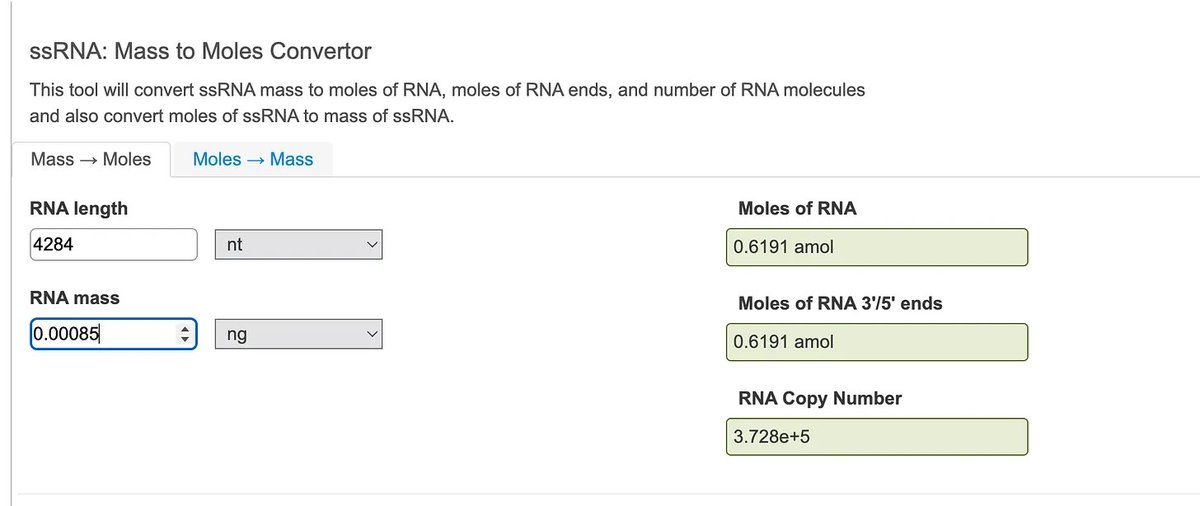
4284bp mRNA at 0.00085ng = 3.728x10^5 molecules.
nebiocalculator.neb.com/#!/ssrnaamt
4284bp mRNA at 0.00085ng = 3.728x10^5 molecules.
nebiocalculator.neb.com/#!/ssrnaamt
@JesslovesMJK @dryostradamus @YoureTheVoiceEF @DJSpeicher @RWMaloneMD @weldeiry @KUPERWASSERLAB @KMilhoanMDPhD @Jikkyleaks @DrJBhattacharya @NicHulscher
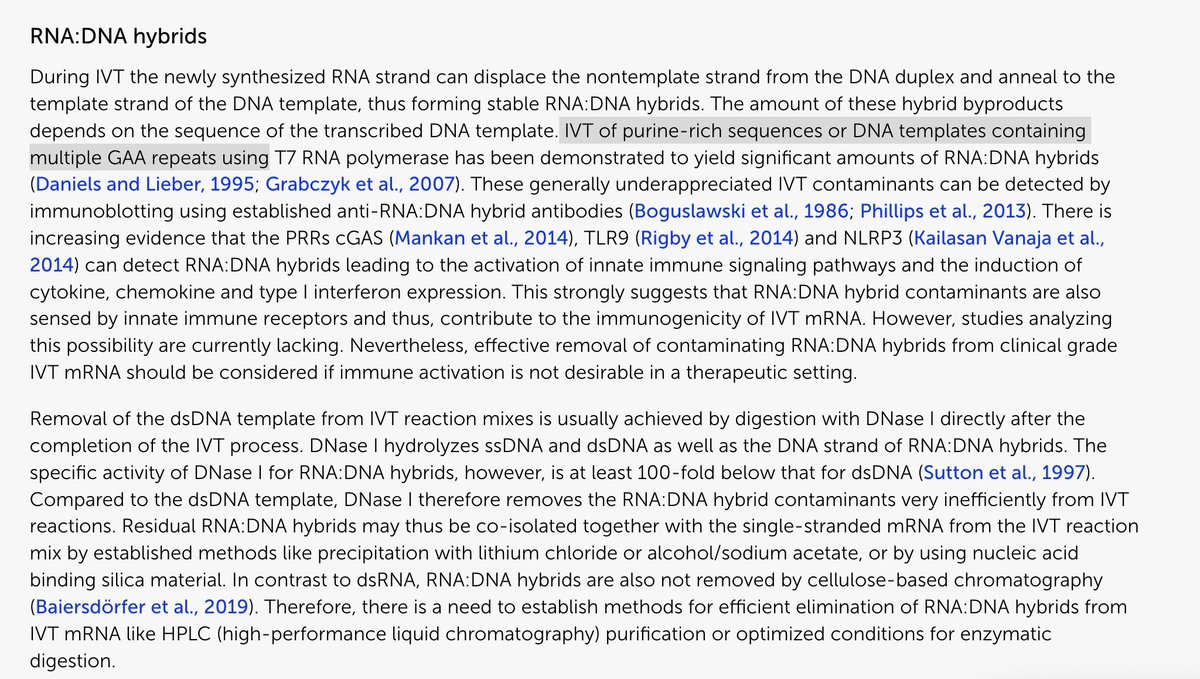
They even try to refute people who measured this before at Yale and Pattersons lab are mistaken as they didnt differentiate C19 from Vax Spike? Thats odd?
open.substack.com/pub/anandamide…
They even try to refute people who measured this before at Yale and Pattersons lab are mistaken as they didnt differentiate C19 from Vax Spike? Thats odd?
open.substack.com/pub/anandamide…
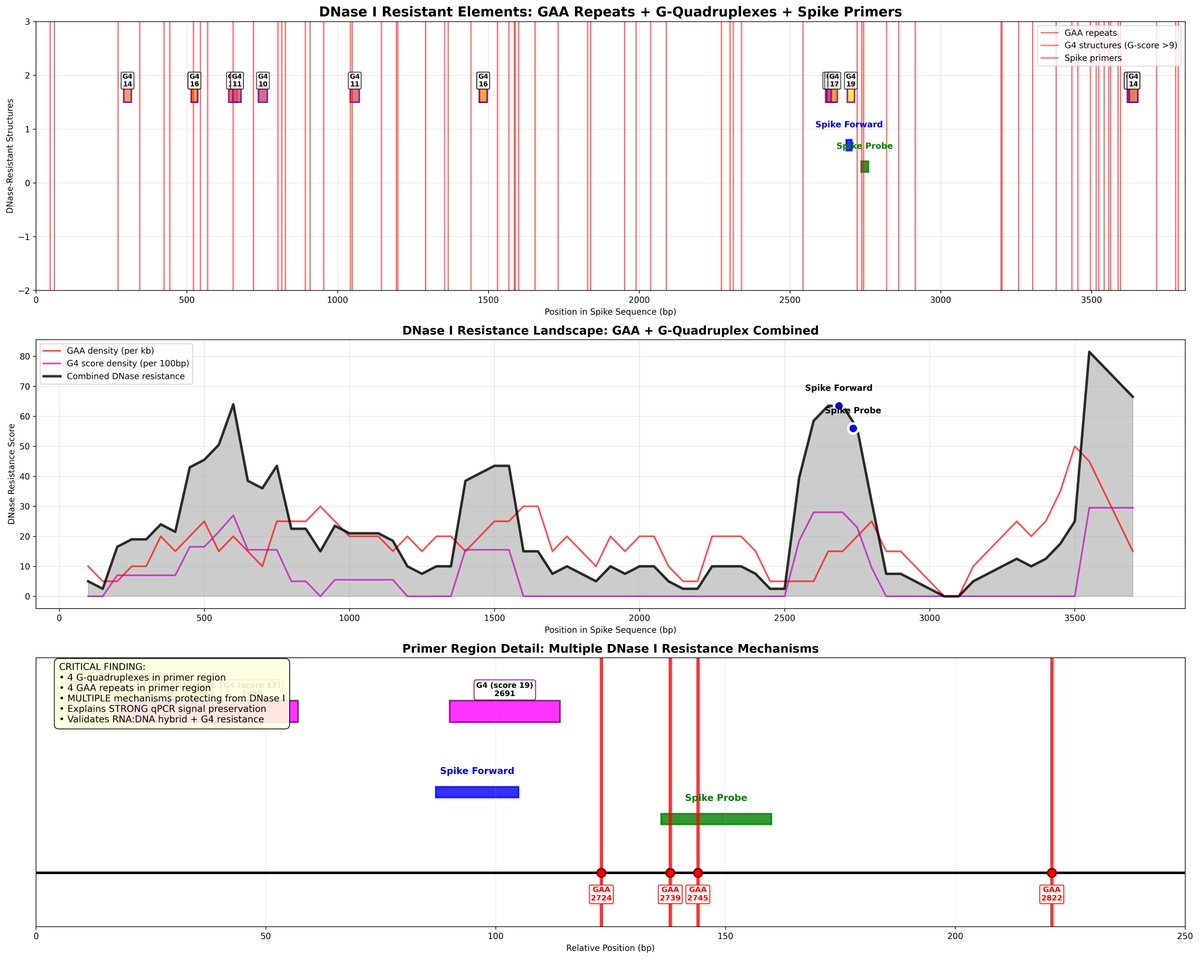
What I find laughable about the paper is that there is no RT-qPCR methods section! No Primers, No cycling conditions, no CT scores, No CT conversion equations, no triplicates, no multiple targets, No 3X dilutions, no Fluorometry or ONT validation?
All things we did in our preprint measuring their vaccines that BioRXIV censored.
All things we did in our preprint measuring their vaccines that BioRXIV censored.
How does @biorxivpreprint explain this bias?
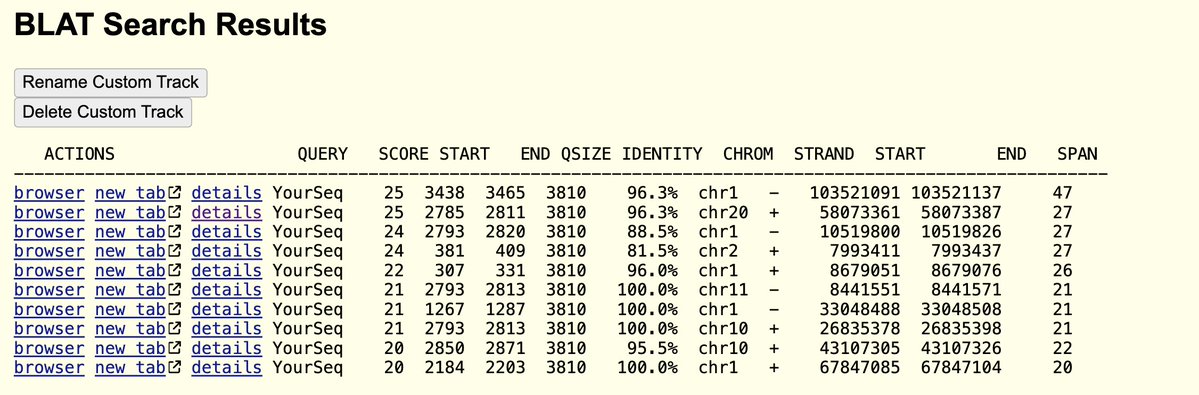
You censor papers using 27 qPCR assays per data point with clear methods sections that eventually get published but let method free papers from Moderna parade around the site?
@SenRonJohnson @RobertKennedyJr @MaryanneDemasi
journalofindependentmedicine.org/articles/v02n0…
You censor papers using 27 qPCR assays per data point with clear methods sections that eventually get published but let method free papers from Moderna parade around the site?
@SenRonJohnson @RobertKennedyJr @MaryanneDemasi
journalofindependentmedicine.org/articles/v02n0…
• • •
Missing some Tweet in this thread? You can try to
force a refresh