Principal Scientist at Altos Labs. Making data-based macromolecular modelling faster, more accurate and a bit more fun. Devoted husband and father.
How to get URL link on X (Twitter) App


 What I'd mostly like to talk about is the saga of how this model came to be... but first, a little about what makes it special. At first glance, it's fairly similar to other bacterial photosystems: a central reaction complex (RC) surrounded by a light-harvesting (LH) ring. (2/)
What I'd mostly like to talk about is the saga of how this model came to be... but first, a little about what makes it special. At first glance, it's fairly similar to other bacterial photosystems: a central reaction complex (RC) surrounded by a light-harvesting (LH) ring. (2/)
https://twitter.com/DavSwain/status/1453095353381707779Remember the big RC-LH1 dimer complex from a few weeks back? These smaller LH2 complexes act as little satellites to that, increasing the light-gathering area and passing their excitation energy back to the LH1 complex. (2/7)
https://twitter.com/CrollTristan/status/1446857820175994889?s=20

https://twitter.com/PDBeurope/status/1418511467989684224There are now 19 structures of this guy in the wwPDB, all post-resolution-revolution cryoEM structures. The first (2017) was reconstructed to a character-building 4.3 Å. I'm told the model building was extremely challenging, ... (2/10)

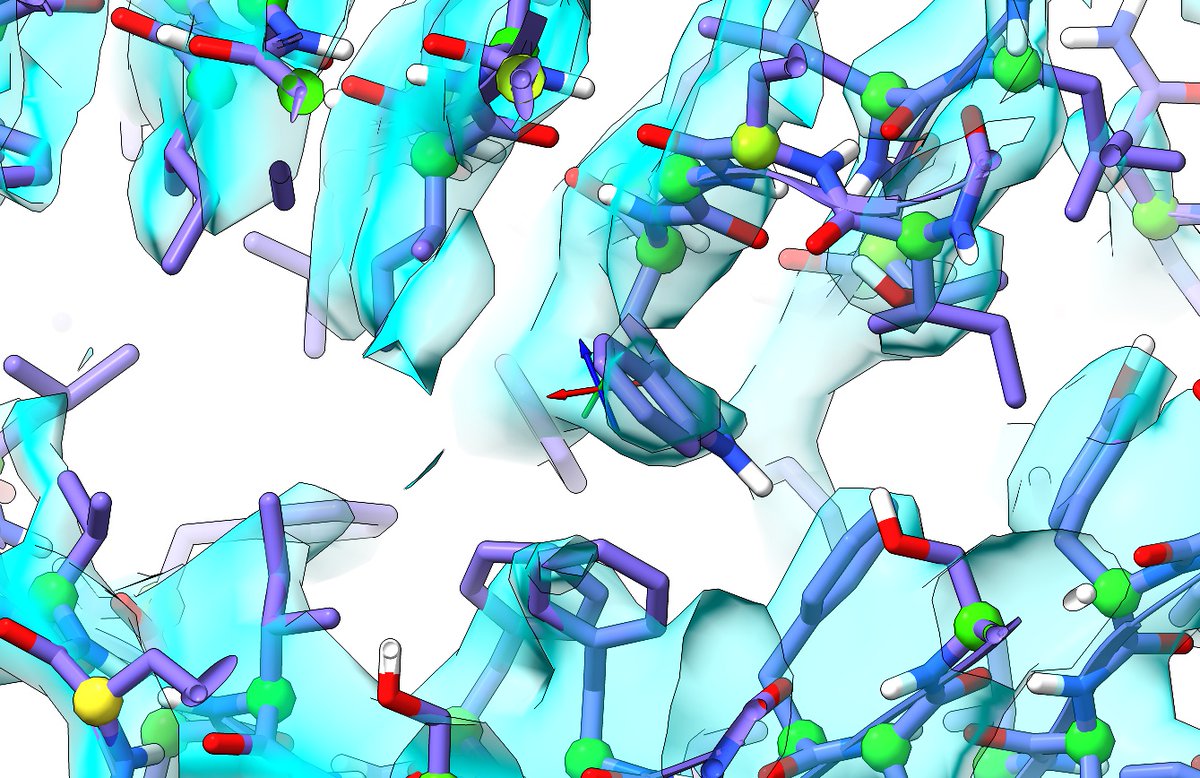
https://twitter.com/CrollTristan/status/1357659731691773953(2~{r},4~{r},5~{r},6~{r})-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-2-[(2~{r},4~{r},5~{r},6~{r})-6-[(1~{r})-1,2-bis(oxidanyl)ethyl]-5-[(2~{s},3~{s},4~{r},5~{r},6~{r})-6-[(1~{s})-1,2-bis(oxidanyl)ethyl]-4-[(2~{r},3~{s},4~{r},5~{s},6~{r})-6-[(1~{s})-2-[(2~{s},3~{s},4~{s},5~{s},6~{r})-...