
Director, Transdivisional Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute | Views are my own.
How to get URL link on X (Twitter) App




https://twitter.com/EricTopol/status/1207745913466179584I (and many others) have been enthusiastic about the idea for years, but if we are to move from promise to reality we need to be careful.

https://twitter.com/genes_pk/status/1205555022081081349?s=21If you know something about biology you can hypothesize about the form of the statistical interaction you expect to see—but (a) you need to know a lot already to do this, and (b) not seeing the expected pattern does not automatically mean your hypothesized mechanism is wrong.

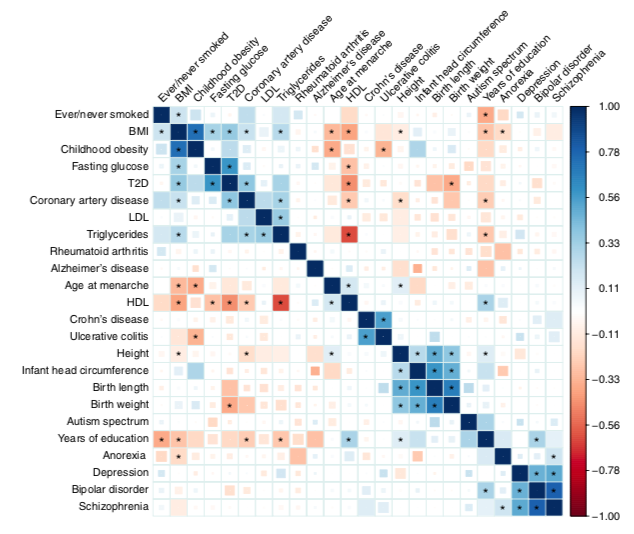

 The question “who—or what—determines populations or groups that merit comparison” is an important but tetchy one. 2/18
The question “who—or what—determines populations or groups that merit comparison” is an important but tetchy one. 2/18

https://twitter.com/GENES_PK/status/1140950476323639296While the potential utility of PRS in any particular context is up for debate, a blanket statement that “GWAS findings are not useful for prediction" is unwarranted.
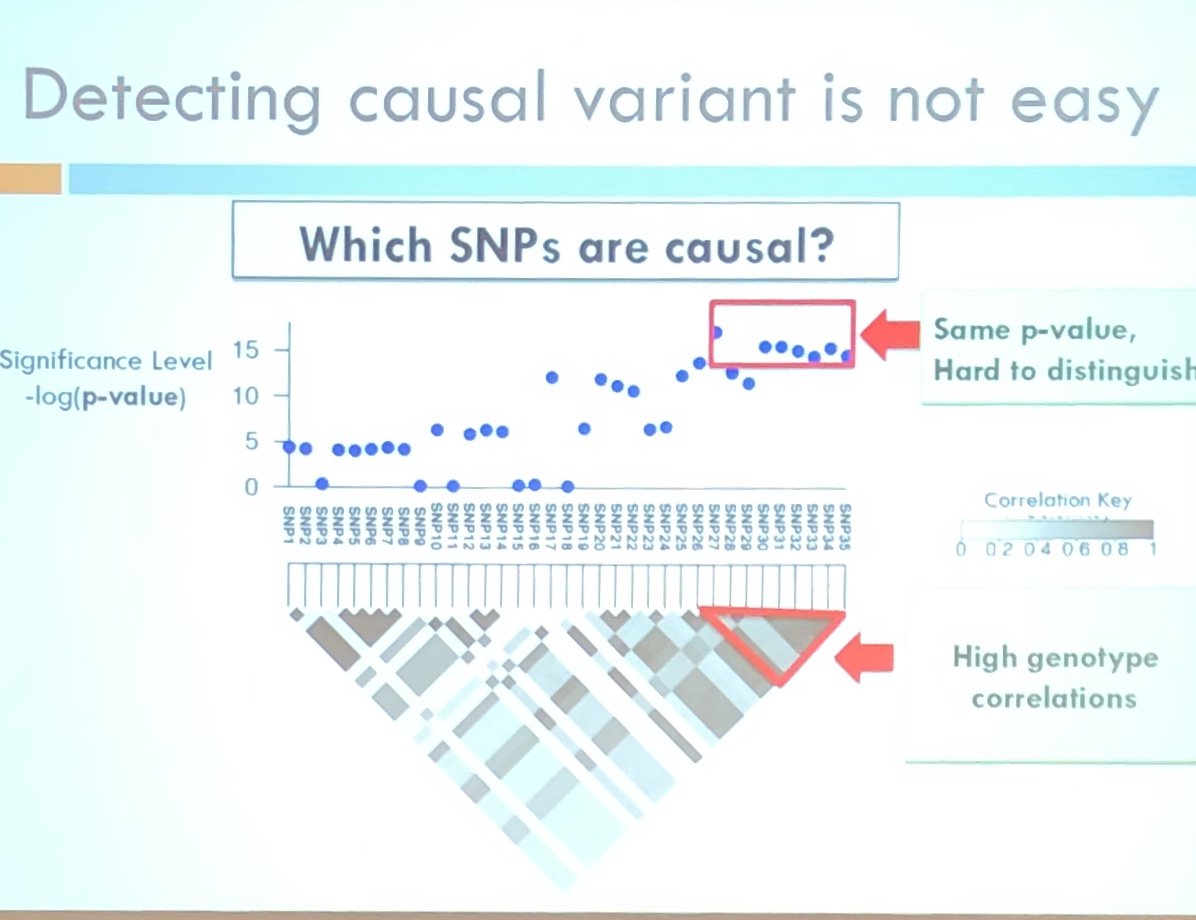
https://twitter.com/venkmurthy/status/1029645513795268608To my mind, the most interesting and important discussion here is given a risk profile similar to that shown in the recent polygenic risk papers—and it need not be a genetic risk factor, could be BMI or circulating plasma rhubarb—what’s the clinical and public health utility? 2/