PhD student @MIT • Research on Generative Models and Geometric Deep Learning for Biophysics • BA @CambridgeUni • Former @TwitterResearch, @DEShawGroup and @IBM
How to get URL link on X (Twitter) App

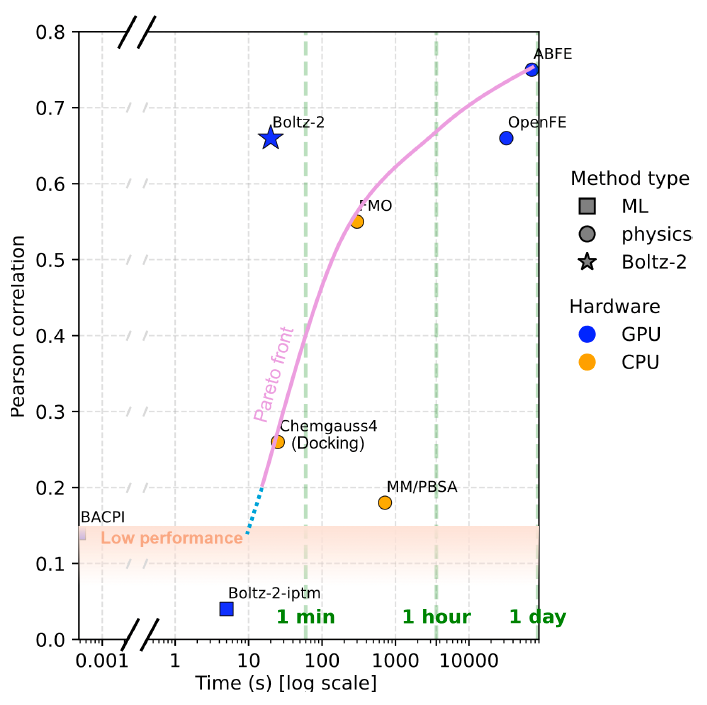
 @NoahBGetz @BarzilayRegina @arkrause TLDR: We studied the problem of flexible molecular docking and the issues with existing methods for the task. We came up with a couple of interesting technical ideas that we validated at small scale in this work and are now making their way into upcoming versions of Boltz! 🔥🚀
@NoahBGetz @BarzilayRegina @arkrause TLDR: We studied the problem of flexible molecular docking and the issues with existing methods for the task. We came up with a couple of interesting technical ideas that we validated at small scale in this work and are now making their way into upcoming versions of Boltz! 🔥🚀

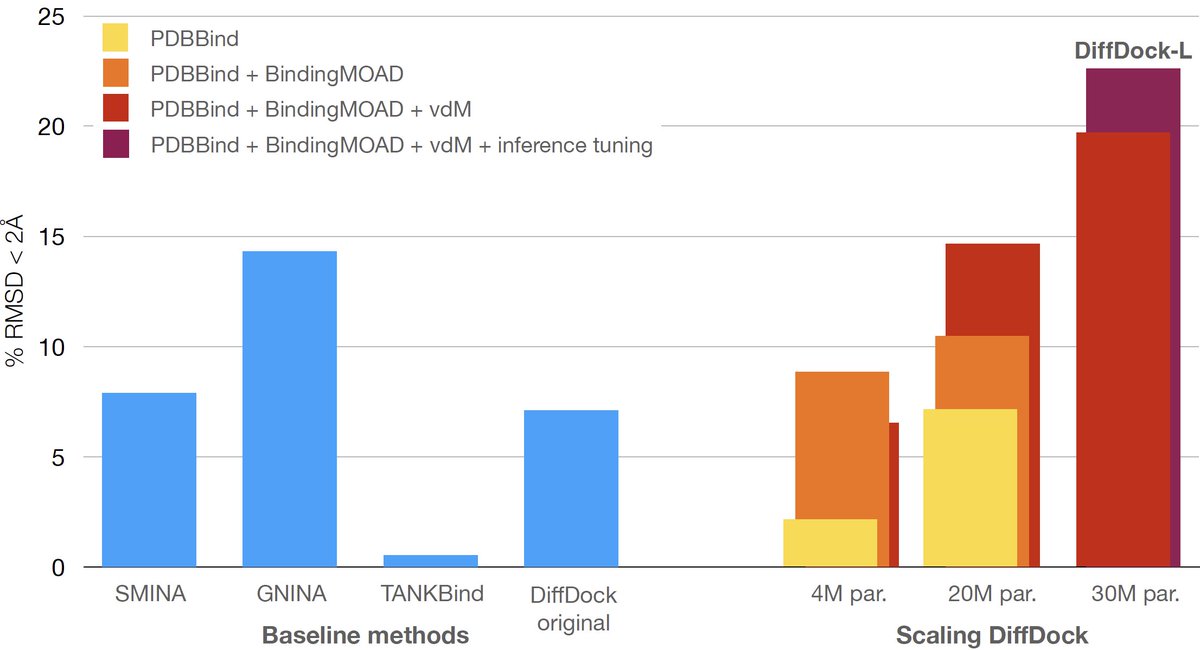
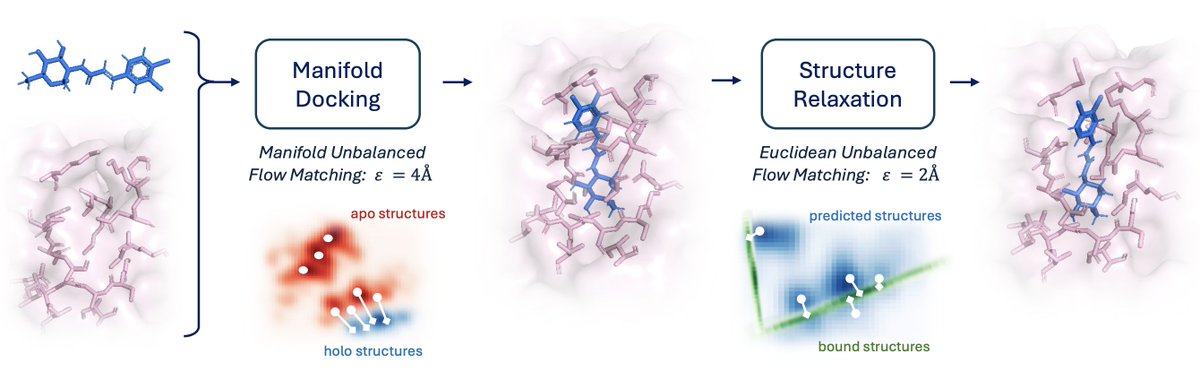
 Solving the general blind docking task would have profound biomedical implications. It would help us understand the mechanism of action of new drugs, predict adverse side-effects before clinical trials… But all these require methods to generalize beyond few well-studied proteins
Solving the general blind docking task would have profound biomedical implications. It would help us understand the mechanism of action of new drugs, predict adverse side-effects before clinical trials… But all these require methods to generalize beyond few well-studied proteins
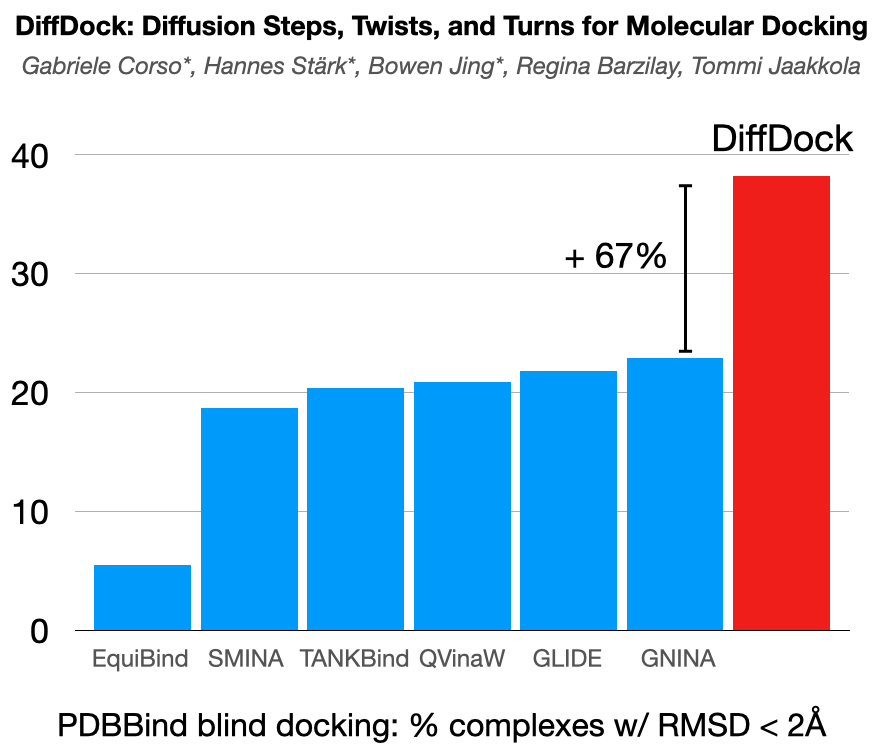
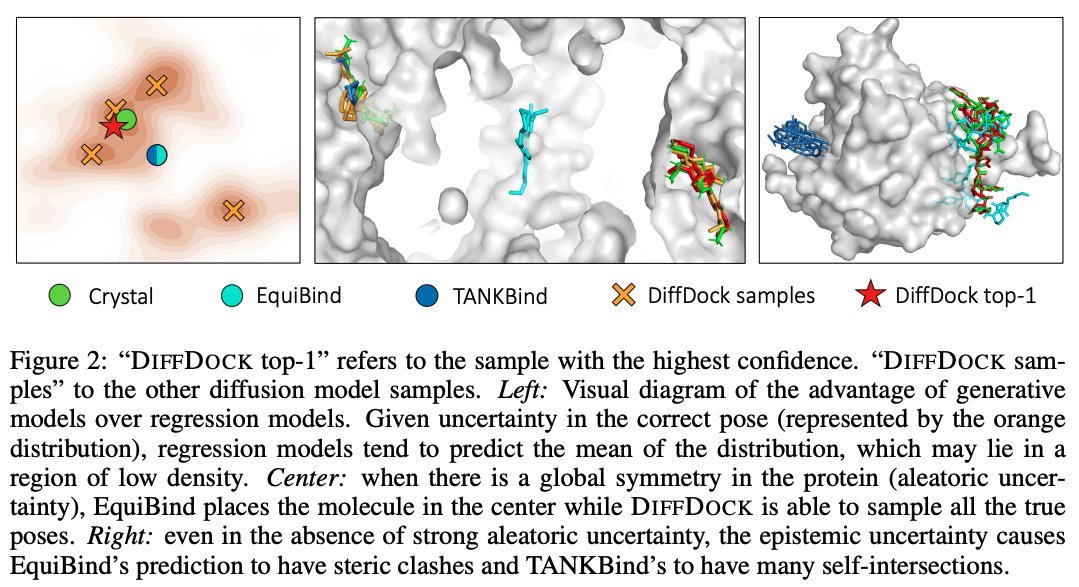
 @HannesStaerk @BarzilayRegina Recent regression-based ML methods for docking showed strong speed-up but no significant accuracy improvements over traditional search-based approaches. We identify the problem in their objective functions and show how generative modeling aligns well with the docking task.
@HannesStaerk @BarzilayRegina Recent regression-based ML methods for docking showed strong speed-up but no significant accuracy improvements over traditional search-based approaches. We identify the problem in their objective functions and show how generative modeling aligns well with the docking task.