
Popgen @UCDavis. @gcbias@ecoevo.social . Tweets, grammar, & spelling are my views only.He/him. #OA popgen book https://t.co/R6I2dcaEGf
How to get URL link on X (Twitter) App











 It's an old trick where folks try to persuade you of the following logic: if you don't buy into hereditarian arguments about behavioural differences among groups, then you must think that genetics makes zero contribution to differences among groups. 2/n
It's an old trick where folks try to persuade you of the following logic: if you don't buy into hereditarian arguments about behavioural differences among groups, then you must think that genetics makes zero contribution to differences among groups. 2/n


 Crossing the west by train is stunning (if tiring)
Crossing the west by train is stunning (if tiring) https://twitter.com/Graham_Coop/status/1418646375022178315


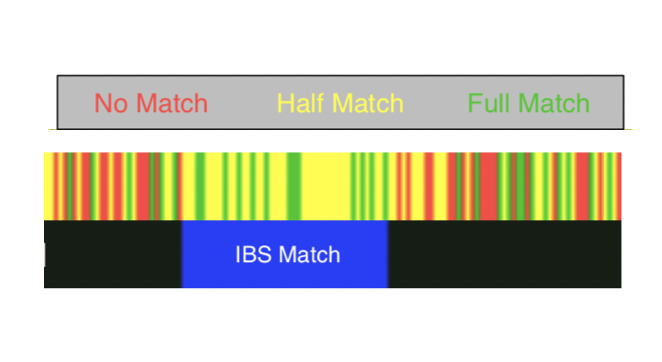


https://twitter.com/jordanmantha/status/1180147511715086338I can claim that the last genealogical common ancestor of humanity lived in Africa or East Asia, and have a similar chance of being right. 2/n

 Original datasets from ideals.illinois.edu/handle/2142/35… (incredible that they have individual-level data from back to 1896). My code here: github.com/cooplab/popgen…
Original datasets from ideals.illinois.edu/handle/2142/35… (incredible that they have individual-level data from back to 1896). My code here: github.com/cooplab/popgen…
 From the papers they reference (biorxiv.org/content/10.110…) they’re constructing these polygenic scores based on LASSO. A standard way to choose a sparse set of SNPs in human genetics
From the papers they reference (biorxiv.org/content/10.110…) they’re constructing these polygenic scores based on LASSO. A standard way to choose a sparse set of SNPs in human genetics