
How to get URL link on X (Twitter) App

 Biology is utterly complex. Luckily, scientists are continuously inventing tools to achieve higher and higher granularity to describe tissues>> cells >> molecules.
Biology is utterly complex. Luckily, scientists are continuously inventing tools to achieve higher and higher granularity to describe tissues>> cells >> molecules.








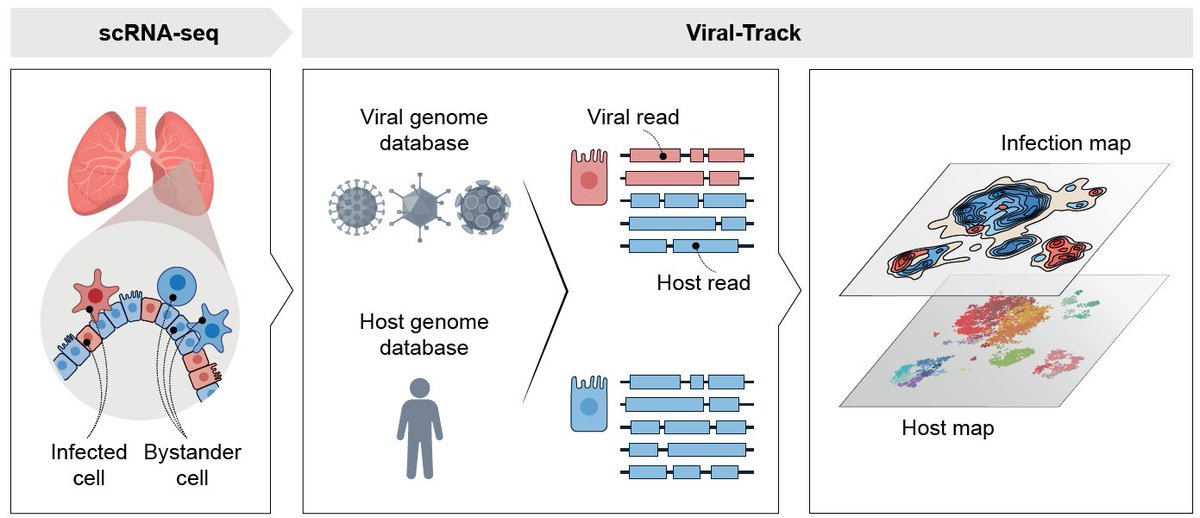


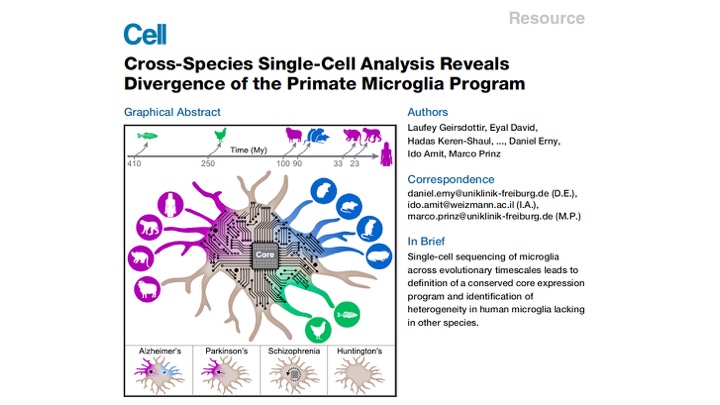
 With the help of our great collaborators, we tracked microglia across 450 million years of evolution by collecting brains from 18 different species, such as blind mole rat, fish, chicken, sheep, monkeys and humans. (2/10)
With the help of our great collaborators, we tracked microglia across 450 million years of evolution by collecting brains from 18 different species, such as blind mole rat, fish, chicken, sheep, monkeys and humans. (2/10) 