
Japanese patent attorney. 🇯🇵🐭 Confronting medical industry that reigns as the power of knowledge. We must not be bystanders to history repeating itself.
4 subscribers
How to get URL link on X (Twitter) App



 The following five Patents 1 to 5 (Patent Publications), each includes the reverse complementary sequence "CTACGTGCCCGCCGAGGAG" in their Sequence Listing.
The following five Patents 1 to 5 (Patent Publications), each includes the reverse complementary sequence "CTACGTGCCCGCCGAGGAG" in their Sequence Listing.
 This FDA document contains multiple sources of information and will be of public interest.
This FDA document contains multiple sources of information and will be of public interest.
 Uğur Şahin, the CEO of BioNTech, has been aware of the problem of the immune imprinting posed by their vaccine at least since February 24, 2023. Despite this, BioNTech did not stop supplying the vaccine and did not deter politicians and experts who recommended booster vaccinations.
Uğur Şahin, the CEO of BioNTech, has been aware of the problem of the immune imprinting posed by their vaccine at least since February 24, 2023. Despite this, BioNTech did not stop supplying the vaccine and did not deter politicians and experts who recommended booster vaccinations.



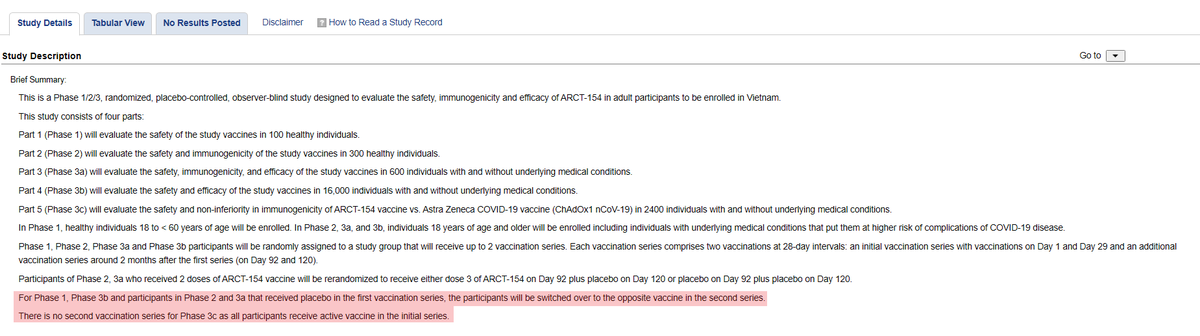 論文では、フェーズ3bで、ワクチン接種者5名、プラセボ接種者16名の計21名が死亡したと報告されています。これは、第1シリーズ後の結果で、第2シリーズの結果は含まれていないようです。
論文では、フェーズ3bで、ワクチン接種者5名、プラセボ接種者16名の計21名が死亡したと報告されています。これは、第1シリーズ後の結果で、第2シリーズの結果は含まれていないようです。





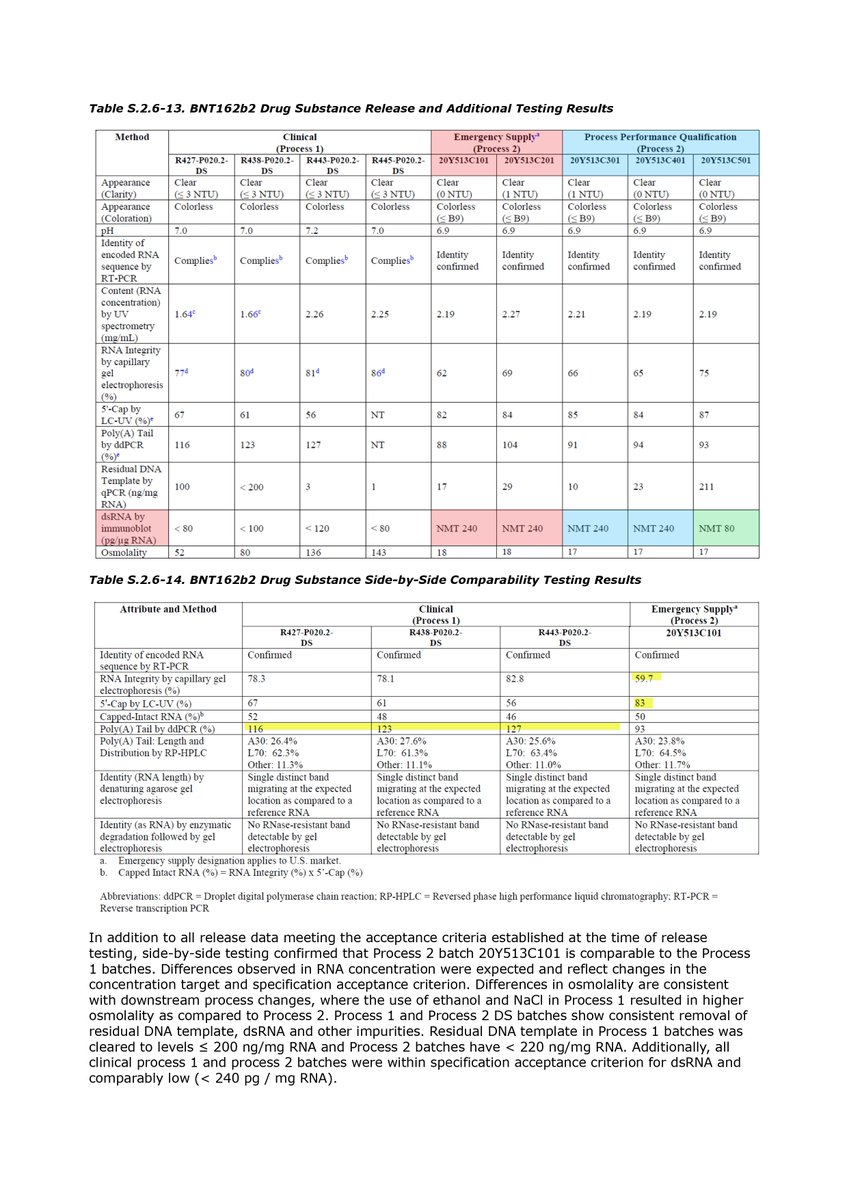 Regarding the relationship between the GTP and UTP concentrations and the dsRNA, the EMA document states as follows:
Regarding the relationship between the GTP and UTP concentrations and the dsRNA, the EMA document states as follows:


 @zombiemommy
@zombiemommy 









 Pfizer (BioNTech) states in the EMA document the benefits of improving the RNA yield and integrity and decreasing the dsDNA by increasing CTP and ATP concentrations. Despite this, Pfizer (BioNTech) concealed the fact that "residual DNA increases" in it.
Pfizer (BioNTech) states in the EMA document the benefits of improving the RNA yield and integrity and decreasing the dsDNA by increasing CTP and ATP concentrations. Despite this, Pfizer (BioNTech) concealed the fact that "residual DNA increases" in it.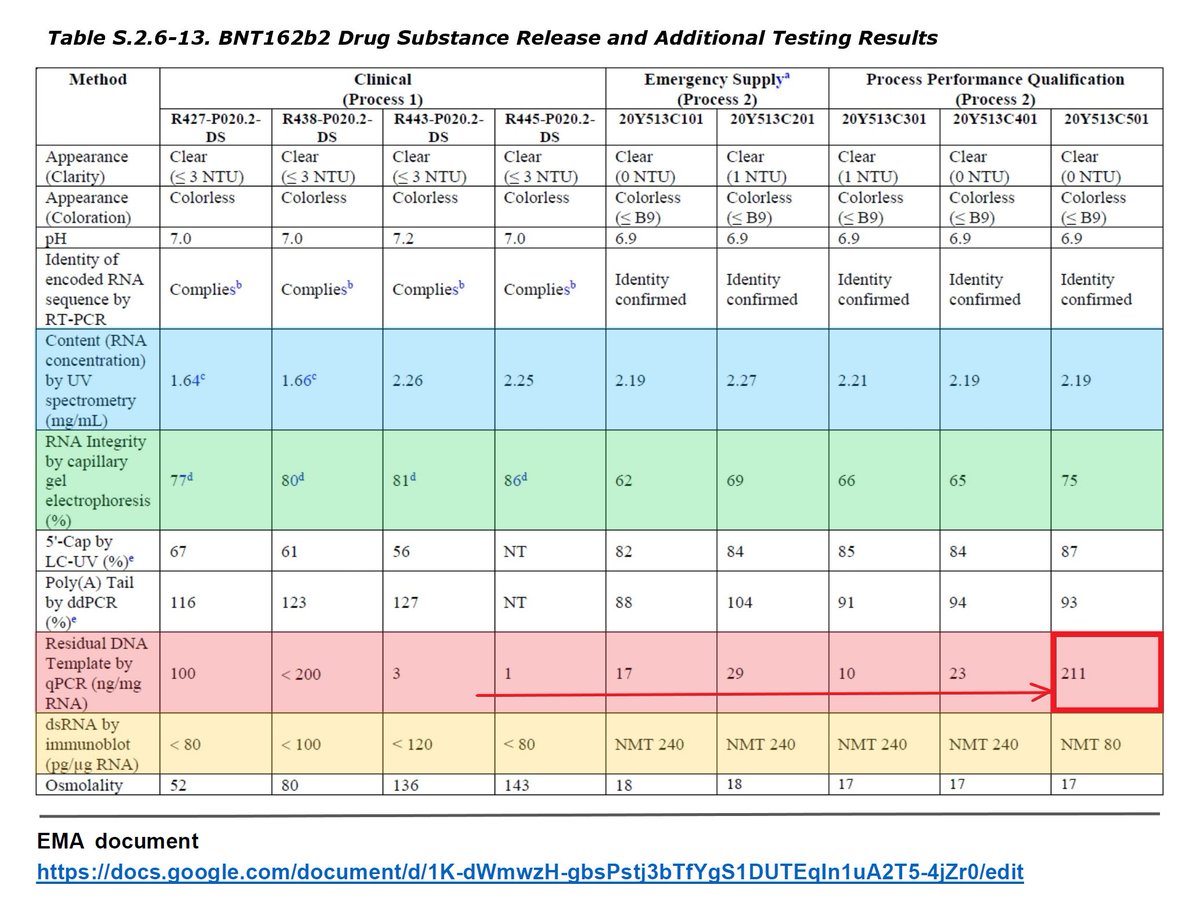

 BioNTech articulates the trade-off between an improving the RNA quality and an increasing the residual DNA in relation to a concentration of CTP (cytidine triphosphate), in the para [0437] and the figure 16 of this patent.
BioNTech articulates the trade-off between an improving the RNA quality and an increasing the residual DNA in relation to a concentration of CTP (cytidine triphosphate), in the para [0437] and the figure 16 of this patent.




 ◆Why is the DNA contamination amount underestimated?
◆Why is the DNA contamination amount underestimated?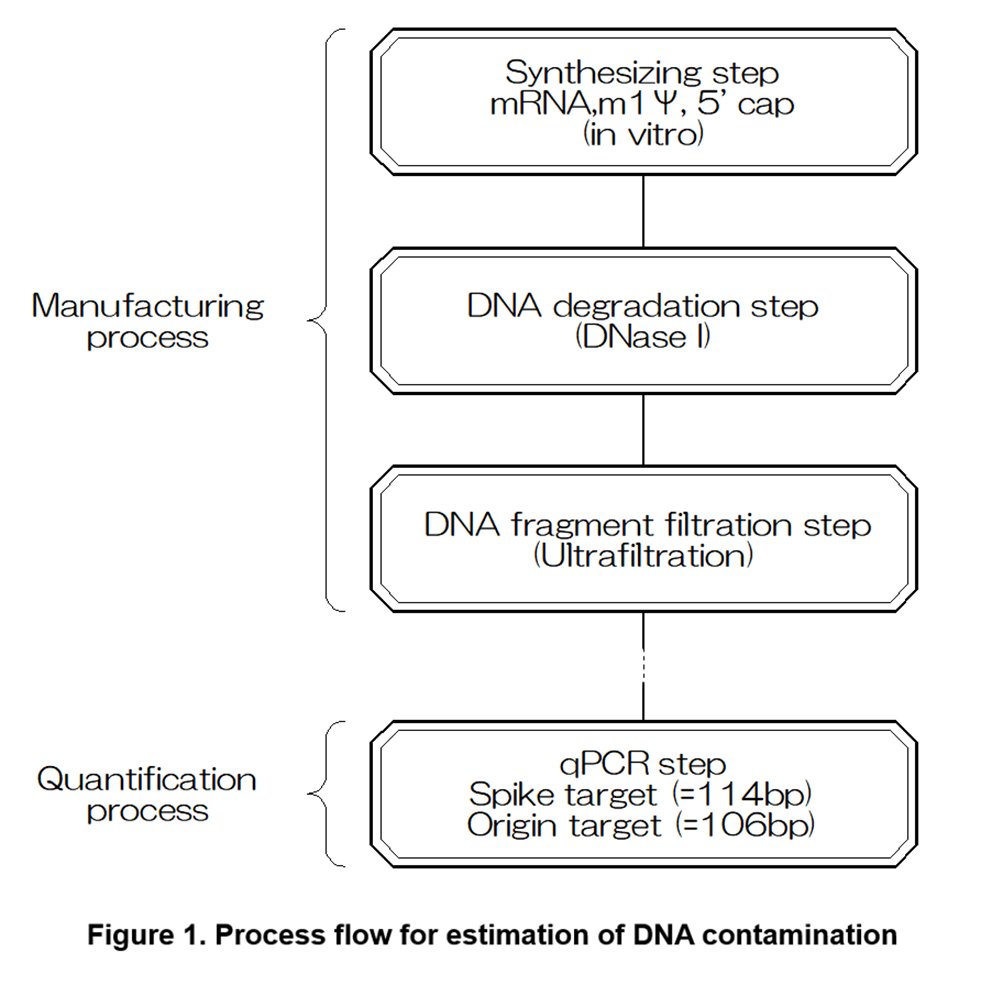



 Further information
Further information https://twitter.com/Patent_SUN/status/1725556303848972410



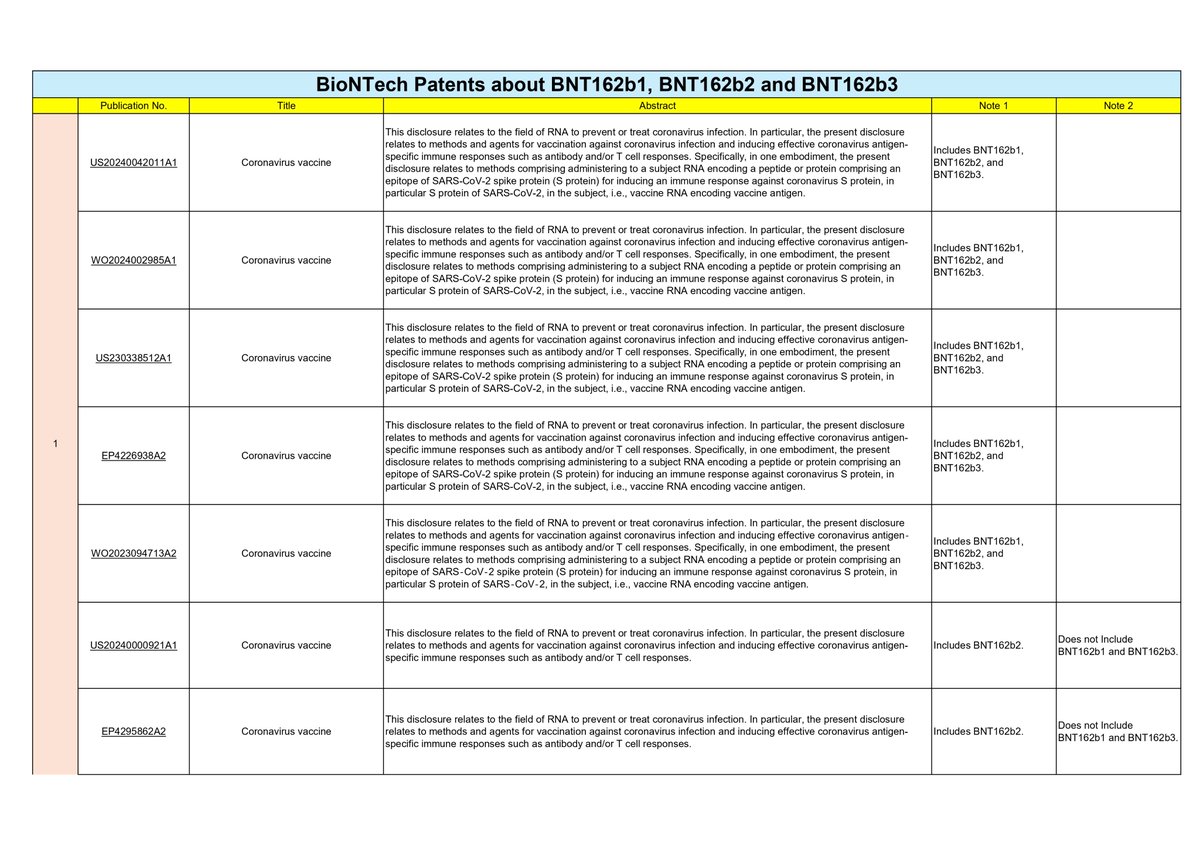
 The Sequence Listing cannot be obtained just by obtaining the patent publications ① to ⑤.
The Sequence Listing cannot be obtained just by obtaining the patent publications ① to ⑤. 



 (1) The spike DNAs
(1) The spike DNAs

https://twitter.com/Kevin_McKernan/status/1682521919612960768?s=20



 Both of the ORF11 region and the ORF19 region have relatively large lengths. The probability of the presence of a DNA fragment including the ORF11 region in an intact state and/or a DNA fragment including the ORF19 region in an intact state therefore may be extremely low.
Both of the ORF11 region and the ORF19 region have relatively large lengths. The probability of the presence of a DNA fragment including the ORF11 region in an intact state and/or a DNA fragment including the ORF19 region in an intact state therefore may be extremely low.







 Both of the ORF11 region and the ORF19 region have relatively large lengths. The probability of the presence of a DNA fragment including the ORF11 region in an intact state and/or a DNA fragment including the ORF19 region in an intact state therefore may be extremely low.
Both of the ORF11 region and the ORF19 region have relatively large lengths. The probability of the presence of a DNA fragment including the ORF11 region in an intact state and/or a DNA fragment including the ORF19 region in an intact state therefore may be extremely low.


