
Former biotech. 📚+🎓= Science, Soc (PH, human rights, int'l law), Psych. RT/studies ≠ endorse/med advice/DX/RX. Sijie Kung Fu. Combat Silat. Views are my own.
35 subscribers
How to get URL link on X (Twitter) App


https://x.com/_HeartofGrace_/status/1965435328585474488
 zenodo.org/records/174449…
zenodo.org/records/174449…
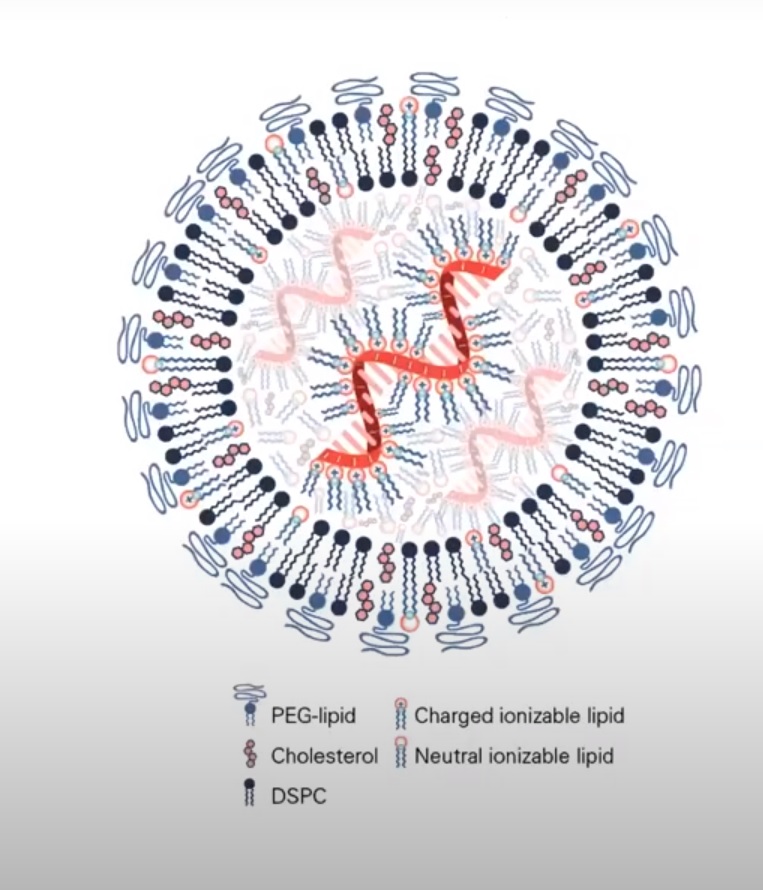

 preprints.org/manuscript/202…
preprints.org/manuscript/202…

 2/ The cGAS-STING Pathway, Ion Channel Dysregulation, and Immune Responses: Implications for Autoimmunity, Inflammation, Long COVID, and Post-Vaccination Responses
2/ The cGAS-STING Pathway, Ion Channel Dysregulation, and Immune Responses: Implications for Autoimmunity, Inflammation, Long COVID, and Post-Vaccination Responses

https://twitter.com/_HeartofGrace_/status/1780785761660383613
 2/
2/ 


 2/ you can read here about the use of a digital twin and cancer.
2/ you can read here about the use of a digital twin and cancer. 

https://twitter.com/Ken__kaneki33/status/1873662544700969202
 2/ Charcot's disease is amyotrophic lateral sclerosis
2/ Charcot's disease is amyotrophic lateral sclerosis 
 2/ Pierre Kory and others have talked a lot about shedding--they are correct. The studies are on viral vectors--these are viral vectors. There are studies I have and so do others on this, but to drive it home--right here:
2/ Pierre Kory and others have talked a lot about shedding--they are correct. The studies are on viral vectors--these are viral vectors. There are studies I have and so do others on this, but to drive it home--right here: 

 2/ . pubmed.ncbi.nlm.nih.gov/36309759/#:~:t…
2/ . pubmed.ncbi.nlm.nih.gov/36309759/#:~:t…

https://twitter.com/_HeartofGrace_/status/1870897973359698347
 2/
2/ 

https://twitter.com/_HeartofGrace_/status/1704615184256335909
 2/ This is an interview and an article done with A recent interview with Dr. Adam Crowe, analytical development manager at Precision NanoSystems Inc.
2/ This is an interview and an article done with A recent interview with Dr. Adam Crowe, analytical development manager at Precision NanoSystems Inc.
https://twitter.com/_HeartofGrace_/status/1869536730656956908
 2/ Looking at this study : "Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva"
2/ Looking at this study : "Fate of free DNA and transformation of the oral bacterium Streptococcus gordonii DL1 by plasmid DNA in human saliva"

 2/🚨 It's true. We are limited on what we have, but to see how plasmid DNA influences promoters on biotech plasmids and plasmid DNA itself is worth paying attention to. Did you tell someone to take zinc who has vaccine injury with suspected DNA plasmid contamination?
2/🚨 It's true. We are limited on what we have, but to see how plasmid DNA influences promoters on biotech plasmids and plasmid DNA itself is worth paying attention to. Did you tell someone to take zinc who has vaccine injury with suspected DNA plasmid contamination?

 2/ osf.io/preprints/osf/…
2/ osf.io/preprints/osf/…

 2/ I went on a podcast two years ago talking about these things and Matt was very gracious and let me talk for over two hours on so many things lipid nanoparticles non stop, it was not even enough time to go over all of this this. Richard and I have talked too about this stuff.
2/ I went on a podcast two years ago talking about these things and Matt was very gracious and let me talk for over two hours on so many things lipid nanoparticles non stop, it was not even enough time to go over all of this this. Richard and I have talked too about this stuff. 

https://twitter.com/EthicalSkeptic/status/1867720731162968066

 2/ Lipid nanoparticle containing RNA,(and DNA contamination in the RNA kind of jabs), lipids, and other KNOWN (yes I said it) and unknown contaminants go all over the body--head to toe. Some cells are more difficult for LNP to get in, like a cardiomyocyte❤️,but they still get in
2/ Lipid nanoparticle containing RNA,(and DNA contamination in the RNA kind of jabs), lipids, and other KNOWN (yes I said it) and unknown contaminants go all over the body--head to toe. Some cells are more difficult for LNP to get in, like a cardiomyocyte❤️,but they still get in 

 x.com/_HeartofGrace_…
x.com/_HeartofGrace_…
https://twitter.com/weldeiry/status/1860320263205749157
 2/ What we would assume if radon was a factor, while not the sole factor, might be an increase in lung cancer occurrence in females under the age of 50 who reside in states with higher levels of radon.
2/ What we would assume if radon was a factor, while not the sole factor, might be an increase in lung cancer occurrence in females under the age of 50 who reside in states with higher levels of radon. 
 2/ "The Effect of Magnetic Field Gradient and Gadolinium-Based MRI Contrast Agent Dotarem on Mouse Macrophages"
2/ "The Effect of Magnetic Field Gradient and Gadolinium-Based MRI Contrast Agent Dotarem on Mouse Macrophages"