
Evolutionary biologist & epidemiologist. COVID-19, antibiotic resistance, cooperation & insect societies. Assistant professor @LSHTM @CMMID_lshtm. He/him.
2 subscribers
How to get URL link on X (Twitter) App


https://twitter.com/BarnardResearch/status/1469646833563615237There was some confusion over what our models, which assumed Omicron had equal "baseline" severity to Delta, meant for severity in practice. This leads to around a 40% reduction in realised severity within each age group, because more Omicron cases are breakthrough/reinfections.


 But before I go on, I will remind my scientific and media readers that SGTF is only a proxy for Omicron, and is also found in other SARS-CoV-2 lineages. Lineage identity can only be confirmed by sequencing, and longer-term surveillance is required to establish any trend.
But before I go on, I will remind my scientific and media readers that SGTF is only a proxy for Omicron, and is also found in other SARS-CoV-2 lineages. Lineage identity can only be confirmed by sequencing, and longer-term surveillance is required to establish any trend.

 The LSHTM team has been analysing B.1.1.7 for signs of increased or decreased severity since late December. We first identified a signal of higher mortality in mid-January. This led to an announcement by the PM and @uksciencechief on 22 January. bbc.co.uk/news/health-55… 2/16
The LSHTM team has been analysing B.1.1.7 for signs of increased or decreased severity since late December. We first identified a signal of higher mortality in mid-January. This led to an announcement by the PM and @uksciencechief on 22 January. bbc.co.uk/news/health-55… 2/16



 We now include new statistical estimates of B.1.1.7 growth, which accord with our earlier model-based estimates—and provide some really interesting plots. (thanks @Alex_Washburne, @inschool4life, @TWenseleers, @seabbs and @sbfnk!) UNDER PEER REVIEW (2/6)
We now include new statistical estimates of B.1.1.7 growth, which accord with our earlier model-based estimates—and provide some really interesting plots. (thanks @Alex_Washburne, @inschool4life, @TWenseleers, @seabbs and @sbfnk!) UNDER PEER REVIEW (2/6) 

 @i_petersen UK scientists originally interpreted this as the variant spreading (slightly) more easily among children than preexisting variants. But with newer data now in, this may have just been a transient effect related to schools being open during the November lockdown. 2/2
@i_petersen UK scientists originally interpreted this as the variant spreading (slightly) more easily among children than preexisting variants. But with newer data now in, this may have just been a transient effect related to schools being open during the November lockdown. 2/2
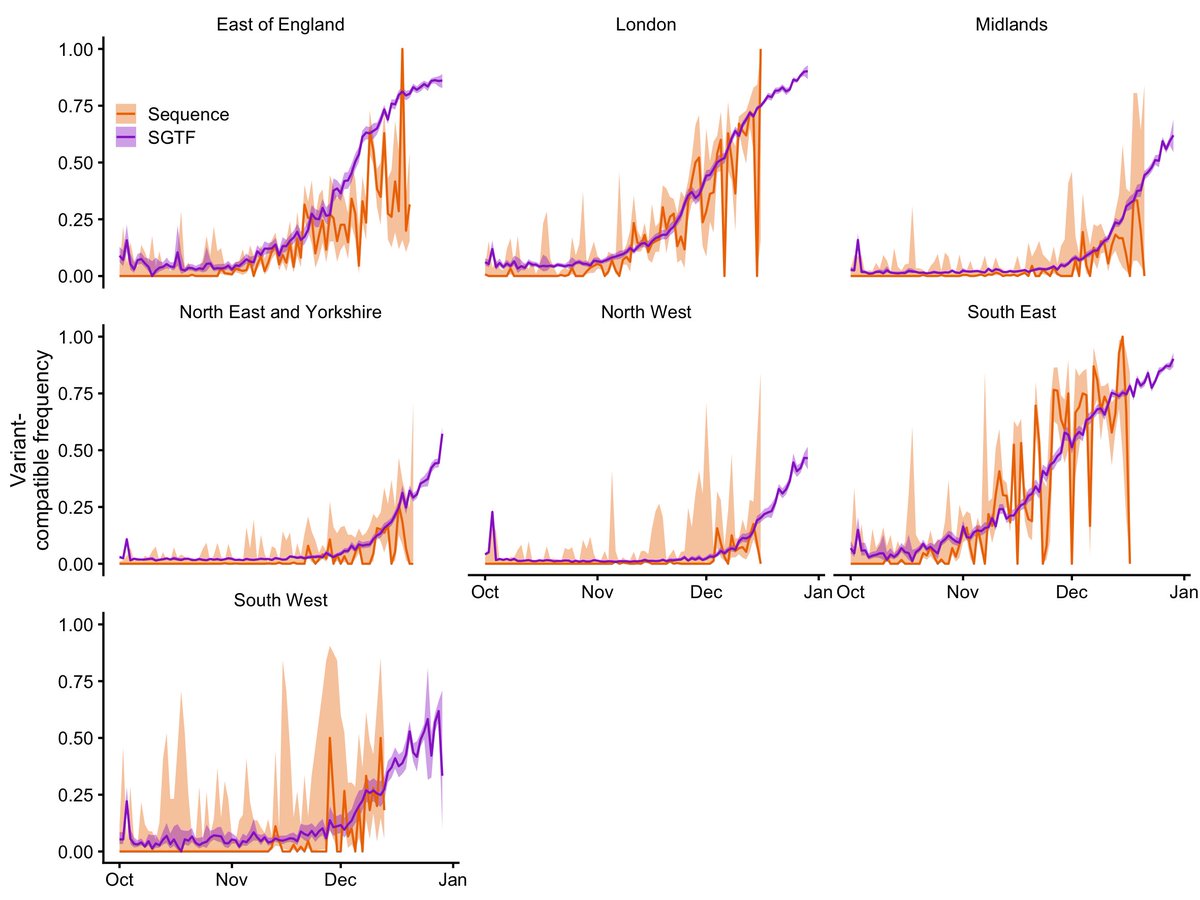
 While the sequencing data above are definitively VOC 202012/01, S-gene target failure (SGTF) is also associated with some other lineages. However, PHE estimates some 98% of SGTF are now VOC 202012/01 (assets.publishing.service.gov.uk/government/upl…). NOT PEER REVIEWED 2/7
While the sequencing data above are definitively VOC 202012/01, S-gene target failure (SGTF) is also associated with some other lineages. However, PHE estimates some 98% of SGTF are now VOC 202012/01 (assets.publishing.service.gov.uk/government/upl…). NOT PEER REVIEWED 2/7

https://twitter.com/_nickdavies/status/1341845176361349123?s=20) Is the apparent spread of VOC due to increased testing? This comes up often when cases rise & indeed case data is subject to biases. But we don't fit to case data in the model. Hospitalisations, deaths, and relative frequency (not abs. number) of the new variant define the trend.
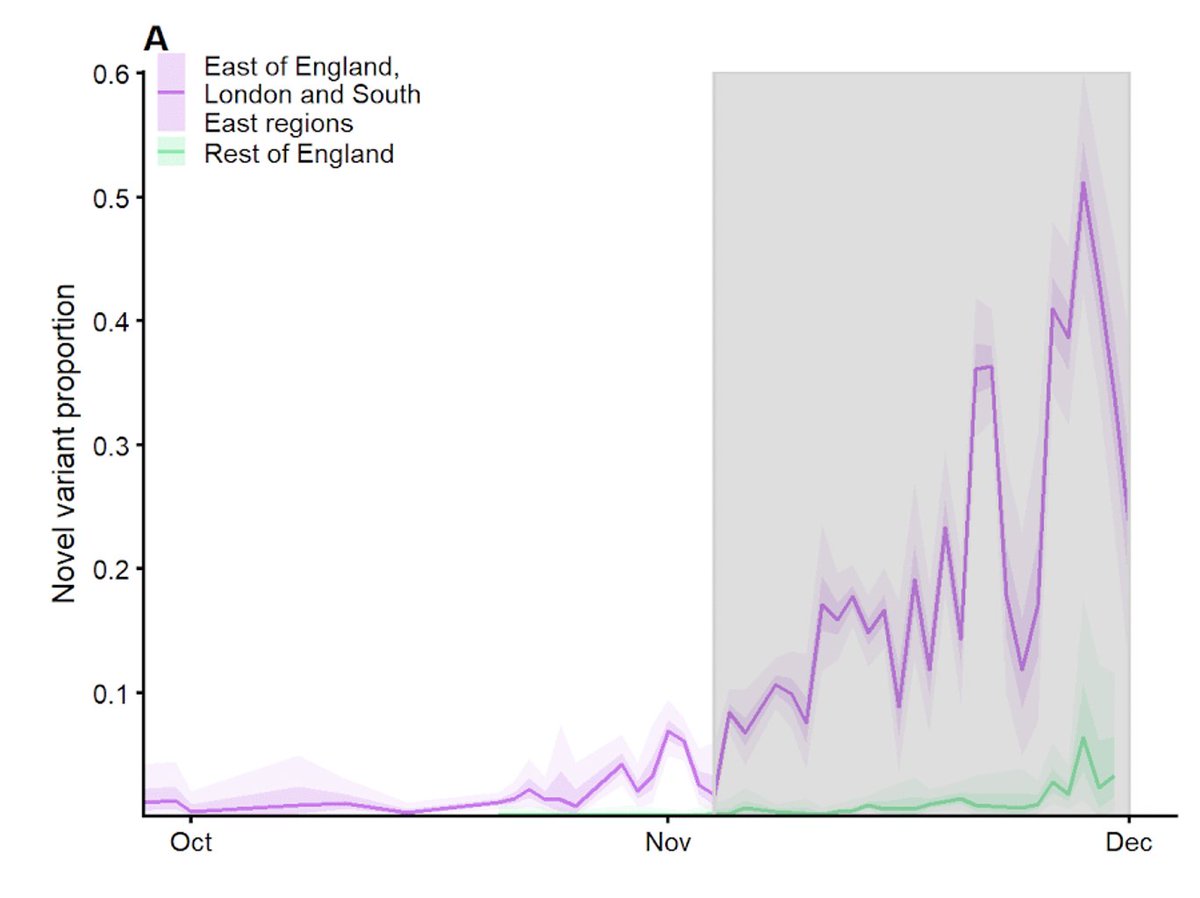
 We fitted a mathematical model to the growth of VOC 202012/01 in these three regions of England. If current trends continue, the new variant could represent 90% of cases by mid-January. NOT PEER REVIEWED 2/9
We fitted a mathematical model to the growth of VOC 202012/01 in these three regions of England. If current trends continue, the new variant could represent 90% of cases by mid-January. NOT PEER REVIEWED 2/9 