
Automating science. Cofounder @EdisonSci. Cofounder @FutureHouseSF. Prof of chem eng @UofR (on sabbatical).
How to get URL link on X (Twitter) App






 link:
link: 
 I was working as a red teamer for GPT-4 and kept getting hallucinated molecules when trying to get up to trouble in chemistry. Then I tried the ReAct agent (from @ShunyuYao12 ) quickly saw real molecules. This work eventually was public in GPT-4 technical report 2/8
I was working as a red teamer for GPT-4 and kept getting hallucinated molecules when trying to get up to trouble in chemistry. Then I tried the ReAct agent (from @ShunyuYao12 ) quickly saw real molecules. This work eventually was public in GPT-4 technical report 2/8 

 Peptide screening usually gives positive examples, which makes it difficult to train a classifier. Previous work has been done on this - including one-class SVM. We evaluate these and propose a modified algorithm built on "spies" 2/4
Peptide screening usually gives positive examples, which makes it difficult to train a classifier. Previous work has been done on this - including one-class SVM. We evaluate these and propose a modified algorithm built on "spies" 2/4

 Bloom filters are fast and can store ultra large chemical libraries in RAM, at the cost of a false positive rate of 0.005 (can tune this!) 2/4
Bloom filters are fast and can store ultra large chemical libraries in RAM, at the cost of a false positive rate of 0.005 (can tune this!) 2/4 

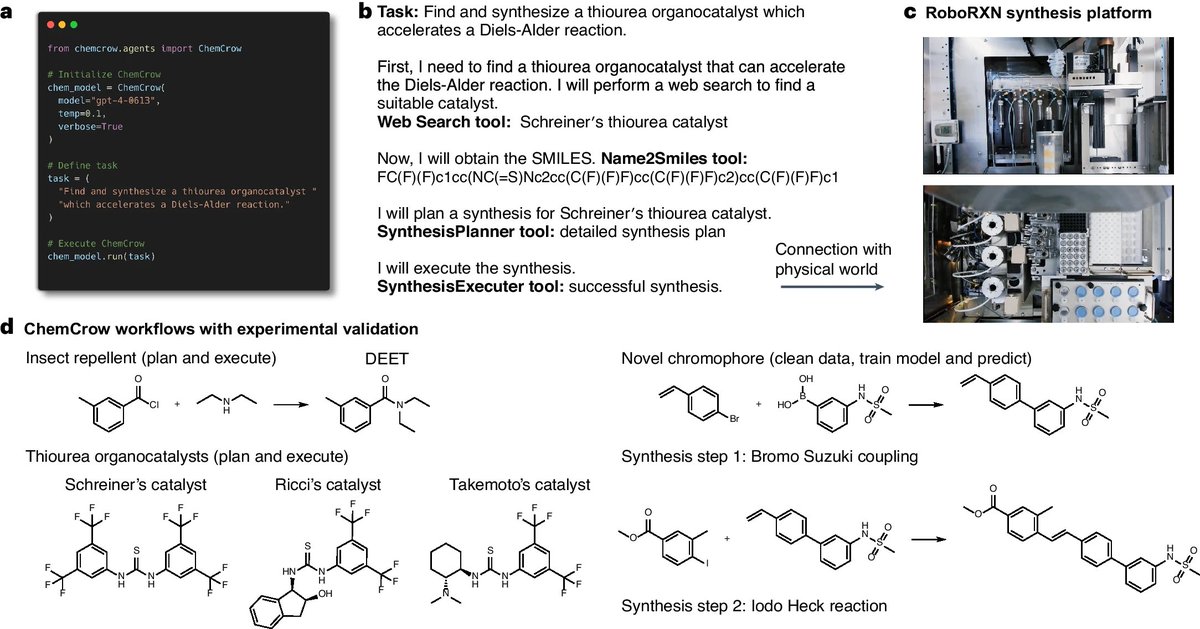
 We, unsurprisingly, found that GPT-4 with tools is much better than GPT-4 alone. Here it outlines a synthesis for atorvastatin complete with steps, an ingredient list, cost, and suppliers. We implement this with @LangChainAI (great library!)
We, unsurprisingly, found that GPT-4 with tools is much better than GPT-4 alone. Here it outlines a synthesis for atorvastatin complete with steps, an ingredient list, cost, and suppliers. We implement this with @LangChainAI (great library!) 

 First - "mutate." Basically create similar molecules from the given molecule. This is interesting in modifying compounds for design or XAI - building out local chemical spaces. 2/4
First - "mutate." Basically create similar molecules from the given molecule. This is interesting in modifying compounds for design or XAI - building out local chemical spaces. 2/4 

 This is one small step in drug discovery. There are many others! The compounds GPT-4 proposes have to be made and tested, and then they just start a path towards a new drug. Let's do a new example for psoriasis by targeting a known protein TYK2. Here is the prompt. 2/n
This is one small step in drug discovery. There are many others! The compounds GPT-4 proposes have to be made and tested, and then they just start a path towards a new drug. Let's do a new example for psoriasis by targeting a known protein TYK2. Here is the prompt. 2/n 




https://twitter.com/PaulRobustelli/status/1455915126566133768


 2/6 LLMs that can generate code have reached accuracy that makes them usable in research. In their training, they picked up knowledge of chemistry. If you ask @OpenAI's Codex to draw caffeine it knows both how to draw a molecule and the structure of caffeine.
2/6 LLMs that can generate code have reached accuracy that makes them usable in research. In their training, they picked up knowledge of chemistry. If you ask @OpenAI's Codex to draw caffeine it knows both how to draw a molecule and the structure of caffeine. 