
Federally funded academic research is the innovation engine of the US economy. Reform is welcome. Destruction will have long term consequences.
How to get URL link on X (Twitter) App

https://twitter.com/pushmeet/status/1937873655888781647Combines several key ideas (a) base-pair resolution profile models (b) sequence context to 1MB (c) unify 1D and 3D tracks (d) simultaneously model multiple readouts across cell contexts (e) model distillation enabling predictive boost of ensembles from a single model. 2/
https://twitter.com/SuvanshSanjeev/status/1935101679856402735These students typically have had access to incredible research opportunities in academia during their undergrads & have spent substantial time doing research already. 2/
https://twitter.com/pdhsu/status/1762512557565456825While incredibly impressive, lets just take the zero-shot gene expression prediction performance of the model (Spearman 𝑟 = 0.41). This is extremely low for a prokaryotic genome 2/
https://twitter.com/SashaGusevPosts/status/1662526641820639234Binarizing signal (eg classifying peaks vs background): please stop this practice for 2 main reasons. Massive loss of info (effect sizes matter), substantial instability of binary models 2/

https://twitter.com/manusaraswat10/status/1528690005576908800A few important caveats (many of which are clearly stated in the paper) (1) this approach is restricted to specific additive architectures, which are often not optimal for different modeling / discovery objectives & data types 2/
https://twitter.com/ewanbirney/status/1019517763122089989And the only way to learn to use NNs is by working with them. Do an actual project with them. Consult with someone familiar with NN to make sure that the problem/dataset is a good match for NNs. 2/
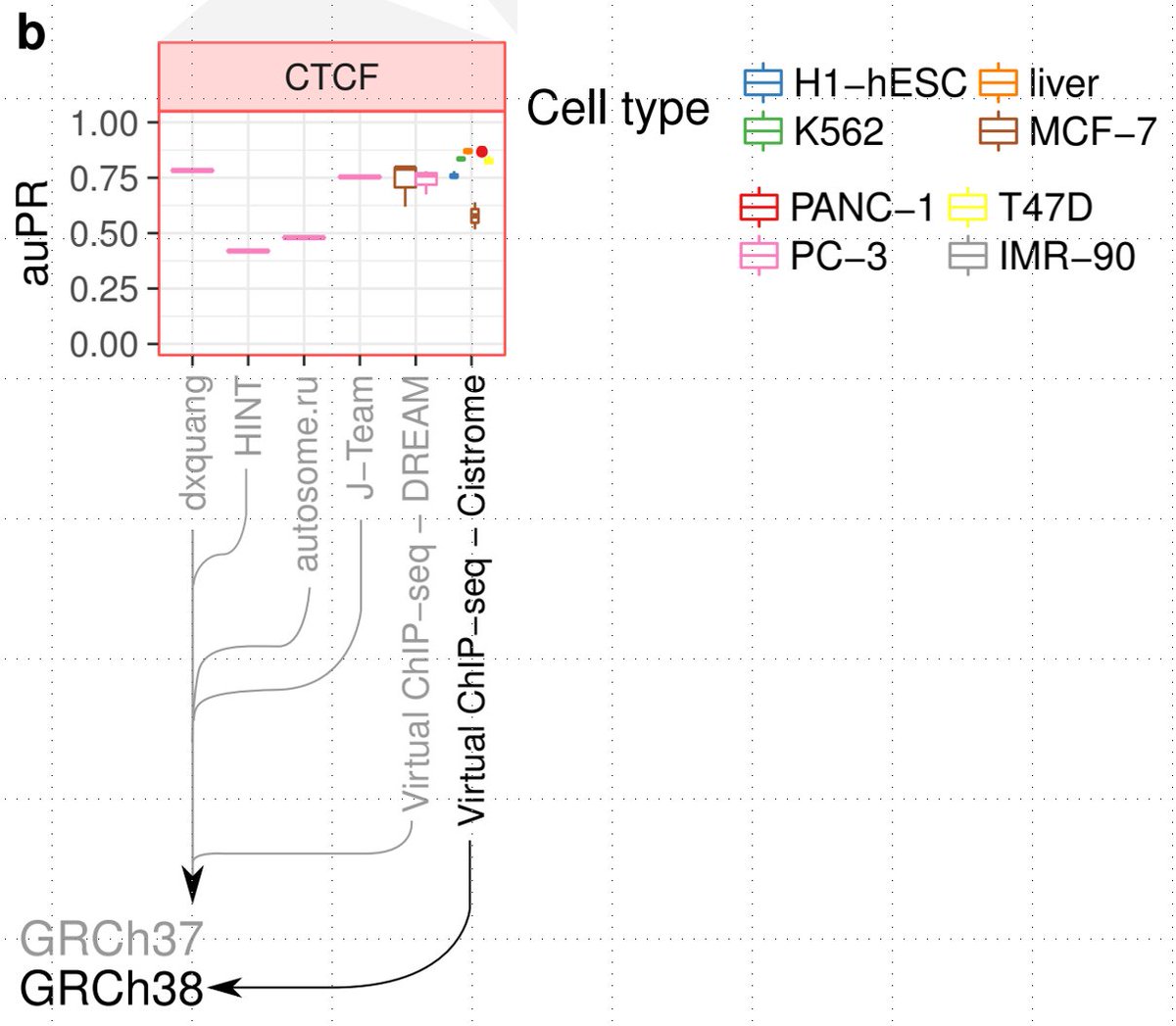
In summary, the paper proposes a powerful imputation based approach for predicting TF binding in NEW cell types. Note that this is the practical prediction problem. Not cross-validation, where u simply hold out chromosomes in cell types where u already have TF ChIP-seq data 1/
External Tweet loading...
If nothing shows, it may have been deleted
by @biorxivpreprint view original on Twitter