
A new scientific institution for curiosity-driven biomedical science and technology.
How to get URL link on X (Twitter) App


 In testing on a 320,000-cell Flex dataset, cyto completed processing in 13 minutes with substantially lower memory and disk usage while producing nearly identical results. Importantly, these gains come from algorithmic efficiency, not just parallelism.
In testing on a 320,000-cell Flex dataset, cyto completed processing in 13 minutes with substantially lower memory and disk usage while producing nearly identical results. Importantly, these gains come from algorithmic efficiency, not just parallelism. 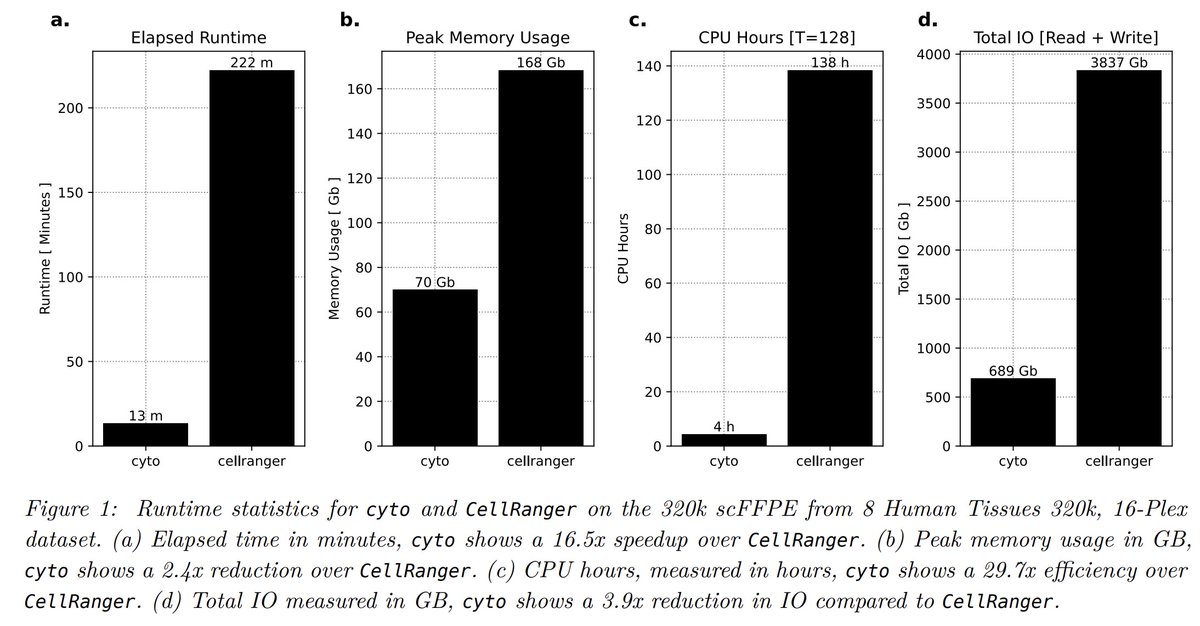
 Stack captures something that most models miss: cellular context. A T cell in inflamed tissue behaves differently, not just because of its own genes, but because of its environment. Stack processes cells together & learns from those relationships.
Stack captures something that most models miss: cellular context. A T cell in inflamed tissue behaves differently, not just because of its own genes, but because of its environment. Stack processes cells together & learns from those relationships. 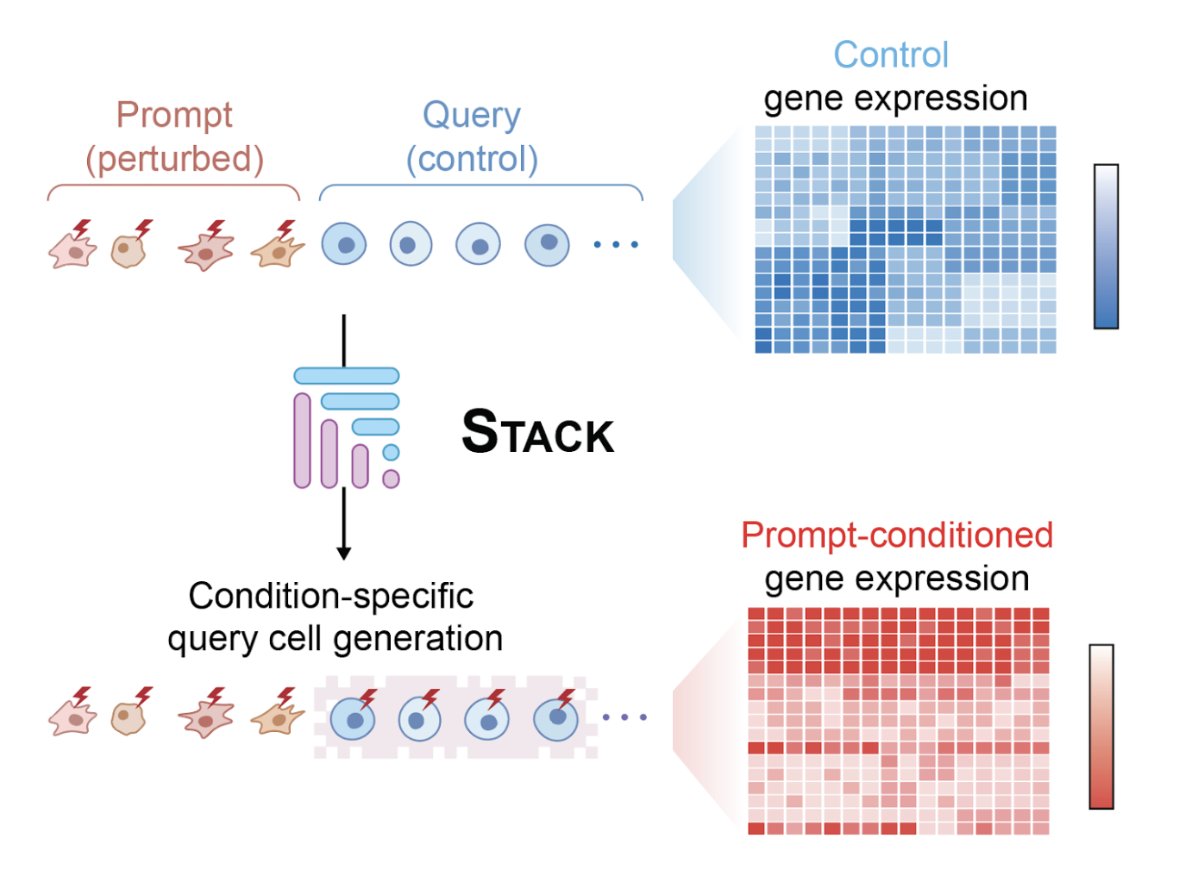
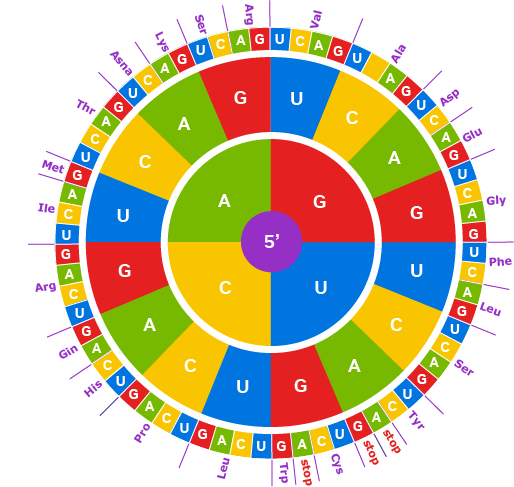


 🪡CRISPR has revolutionized gene editing. But for true large-scale genome design, scientists need a precise and programmable way to rearrange large segments of DNA. The bridge recombinase - discovered in mobile genetic elements - joins any two DNA molecules in a modular way.
🪡CRISPR has revolutionized gene editing. But for true large-scale genome design, scientists need a precise and programmable way to rearrange large segments of DNA. The bridge recombinase - discovered in mobile genetic elements - joins any two DNA molecules in a modular way.