@huggingface engineer. I'm the reason your LLM frontend has a jinja2cpp dependency. Sometimes yells about housing and trans rights instead of working
He/him
8 subscribers
How to get URL link on X (Twitter) App


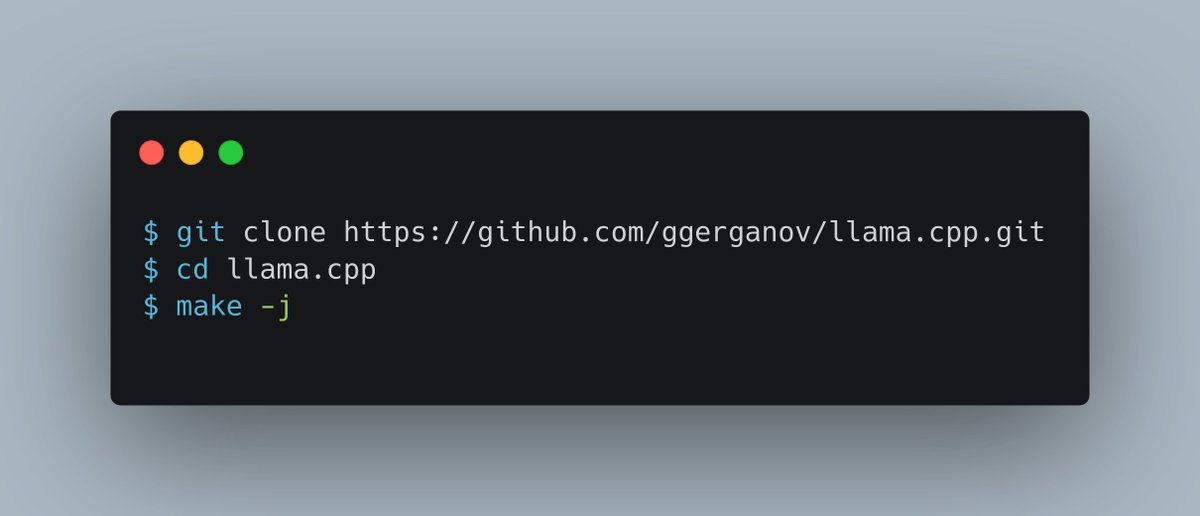
 Tool use with LLMs is one of those things that's simple in theory but surprisingly complex in practice. When the model calls a tool, how do you know? How do you add it to the chat? Did you know some models expected tool defs in JSON schema and others expected Python headers?
Tool use with LLMs is one of those things that's simple in theory but surprisingly complex in practice. When the model calls a tool, how do you know? How do you add it to the chat? Did you know some models expected tool defs in JSON schema and others expected Python headers?