
Genomics, big data, open science. Director of the Centre for Population Genomics at @GarvanInstitute and @MCRI_for_kids. Opinions entirely my own.
How to get URL link on X (Twitter) App




https://twitter.com/ashleyruba/status/1485708682583281666Academics spend their formative years surrounded by other academics. When your mentors, your peers, and the people you most admire have all chosen the same career path, it's hard not to see that as *the* model of success.
https://twitter.com/AprilPawluk/status/1486327770204545036

 We’re incredibly fortunate to be in a position to keep our kids home. My heart goes out to the many parents who can’t - and I’m absolutely furious at the NSW government for not imposing basic common sense safety measures.
We’re incredibly fortunate to be in a position to keep our kids home. My heart goes out to the many parents who can’t - and I’m absolutely furious at the NSW government for not imposing basic common sense safety measures.
https://twitter.com/konradjk/status/1179416576312954880?s=20The error mode results from the presence of low-level contamination in some gnomAD samples, resulting in a small number of reference reads at homozygous non-reference sites; in some cases, GATK will falsely interpret these as supporting a heterozygous genotype.
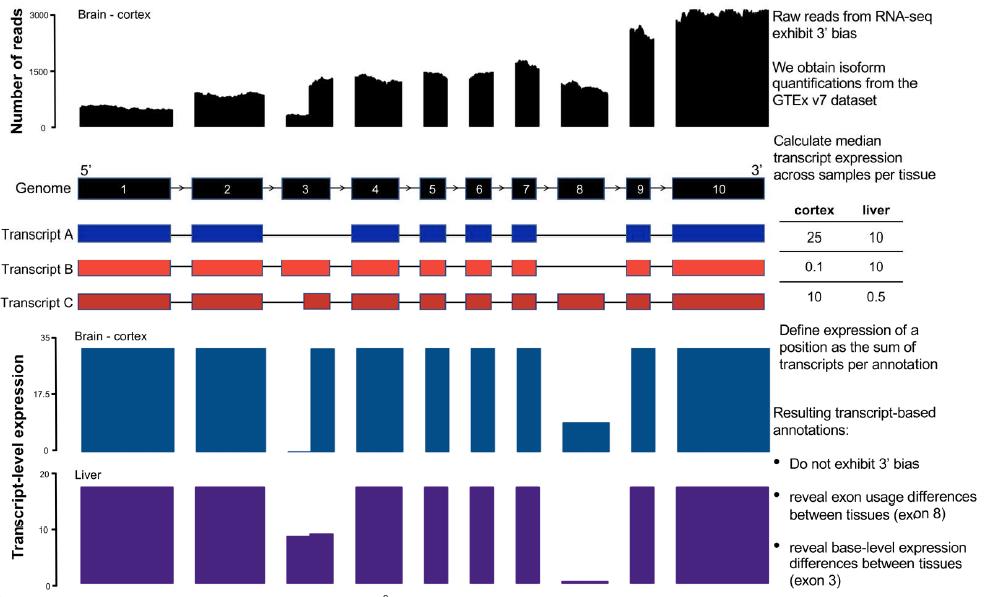
https://twitter.com/beryl_bbc/status/1097920049564798976Our team manually curated 401 of these variants, and found (as expected) a ton of errors. One of the major error types was what we called a "transcript error", meaning the variant fell on a transcript that looked like it wasn't real, or was very weakly expressed.