
Bren Professor of Computational Biology @caltech. Blog at https://t.co/FFQzhEsmhi. Tweets represent my views, not my employer's. #methodsmatter
4 subscribers
How to get URL link on X (Twitter) App







https://twitter.com/SnyderShot/status/1823814971761025451The main result about major changes in the mid 40s and 60s is shown in this plot (Fig. 4a). First, I redrew it with axes that start at 0, so the scale of change here was clearer. Not as impressive, but maybe it's a thing? 2/



https://x.com/lpachter/status/1814716374847197354At issue are benchmarking results we performed comparing our tool, lr-kallisto, to other programs including Patro's Oarfish. Shortly after we posted our preprint Patro started subtweeting our work, claiming we'd run an "appallingly wrong benchmark" and that we're "bullies". 2/


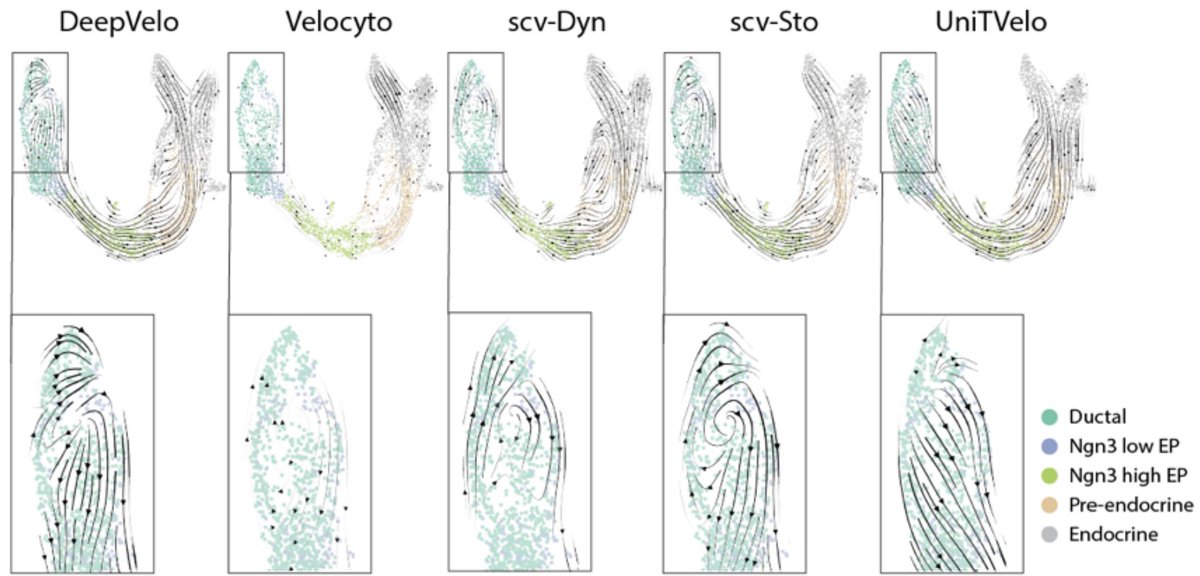
 This is of course no surprise. In "RNA velocity unraveled" by @GorinGennady et al. in @PLOSCompBiol we wrote 55 page paper explaining the many ways in which RNA velocity makes no sense. 2/ journals.plos.org/ploscompbiol/a…
This is of course no surprise. In "RNA velocity unraveled" by @GorinGennady et al. in @PLOSCompBiol we wrote 55 page paper explaining the many ways in which RNA velocity makes no sense. 2/ journals.plos.org/ploscompbiol/a…

https://twitter.com/anshulkundaje/status/1816590665993818292First, the claim that "lower OPC fraction across regions and, in particular, in non-neocortex regions was significantly associated with impaired cognition (Supplementary Fig. 37d)" is not true. Supp. Fig. 37d is below. I've boxed in red the panel the claim is based on. 2/






 The input to splitcode are reads in FASTQ, along with a config file. The output can include edited reads or extracted subsequences, in FASTQ (including gzipped), BAM, or interleaved sequences to stdout. Regions can be identified using absolute location or relative anchors. 2/
The input to splitcode are reads in FASTQ, along with a config file. The output can include edited reads or extracted subsequences, in FASTQ (including gzipped), BAM, or interleaved sequences to stdout. Regions can be identified using absolute location or relative anchors. 2/ 

https://twitter.com/lpachter/status/1787043697541992692Sosúa is a beautiful place in Puerto Plata on the north coast of the Dominican Republic. About 56,000 people live there now.



https://twitter.com/lpachter/status/1776280345098494025The change was described in release notes on May 17, 2022, which via two clicks lead to a technical note with more detail: 2/ cdn.10xgenomics.com/image/upload/v…

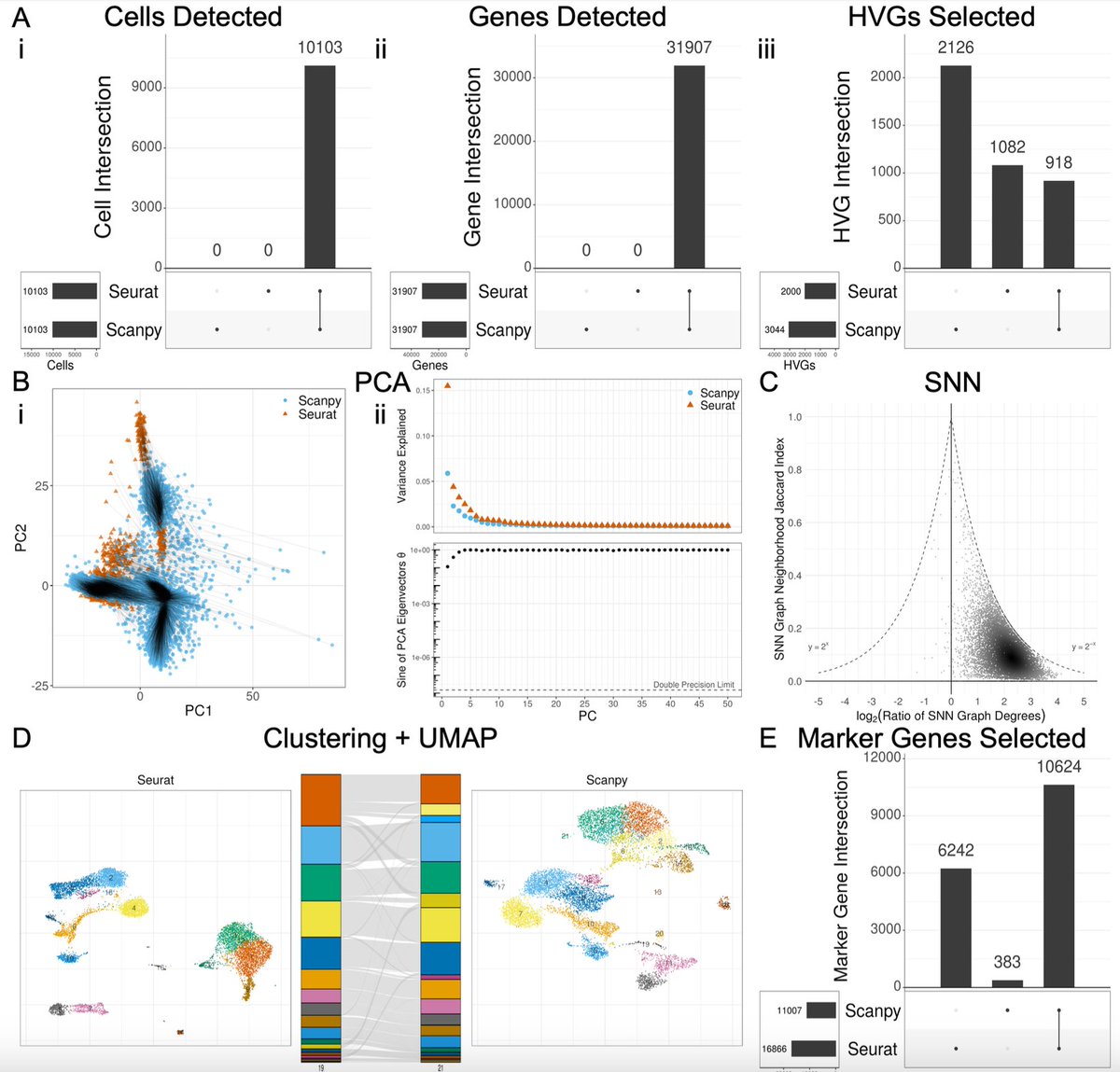
 We looked at a standard processing / analysis summarized in the figure below. The sources of variability we explored are in red. The plots and metrics we assessed are in blue. We examined the standard benchmark 10x PBMC datasets, but results can be obtained for other data. 2/
We looked at a standard processing / analysis summarized in the figure below. The sources of variability we explored are in red. The plots and metrics we assessed are in blue. We examined the standard benchmark 10x PBMC datasets, but results can be obtained for other data. 2/ 

 Just today I revisited the PCA post to recall some of the properties of the transform. A student, Nick Markarian, taught me the Borel-Kolmogorov paradox today (topic for a future post) and the post was helpful in thinking about some things. 2/ liorpachter.wordpress.com/2014/05/26/wha…
Just today I revisited the PCA post to recall some of the properties of the transform. A student, Nick Markarian, taught me the Borel-Kolmogorov paradox today (topic for a future post) and the post was helpful in thinking about some things. 2/ liorpachter.wordpress.com/2014/05/26/wha…

https://x.com/dvir_a/status/1648420062062329856The topic came up at dinner. History presents a heavy burden for Jews in Bonn.. even 78 years after WWII. The "Hans in luck" restaurant we were dining at is just a few meters from where the local synagogue was burned down during "Kirstallnacht" in 1938. 2/


https://x.com/mcnees/status/1418572574947217419?s=20

https://twitter.com/FareedZakaria/status/1733927077085143263The speech begins with the claim that the public has been losing faith in universities because universities are pushing political agendas instead of catering to excellence. But he provides no evidence of a link.

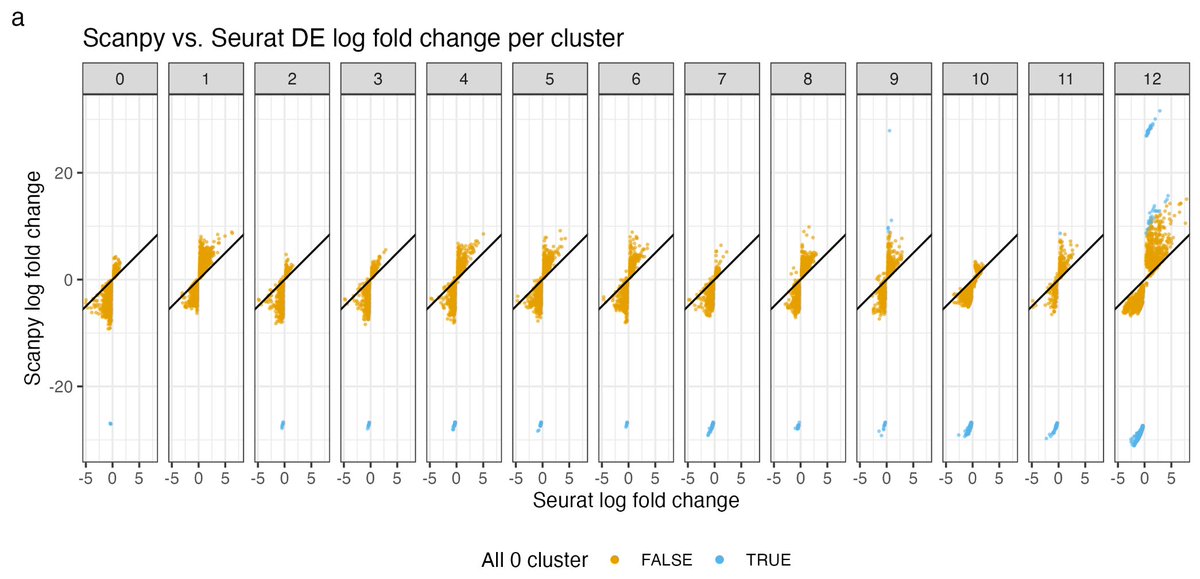
https://twitter.com/lpachter/status/1693880427948421193
 I first became aware of the discrepancy in LFC reporting by Seurat and Scanpy from a preprint by @jeffreypullin and @davisjmcc:
I first became aware of the discrepancy in LFC reporting by Seurat and Scanpy from a preprint by @jeffreypullin and @davisjmcc: https://twitter.com/slavov_n/status/1582347828818456576
https://twitter.com/const_ae/status/1645455772468273153The "performance" in this analysis boils down to checking consistency of the kNN graph after transformation. That's certainly a property one can optimize for, but it's by no means the only one. In fact, if it was the only property of interest, one could just not transform. 2/