How to get URL link on X (Twitter) App

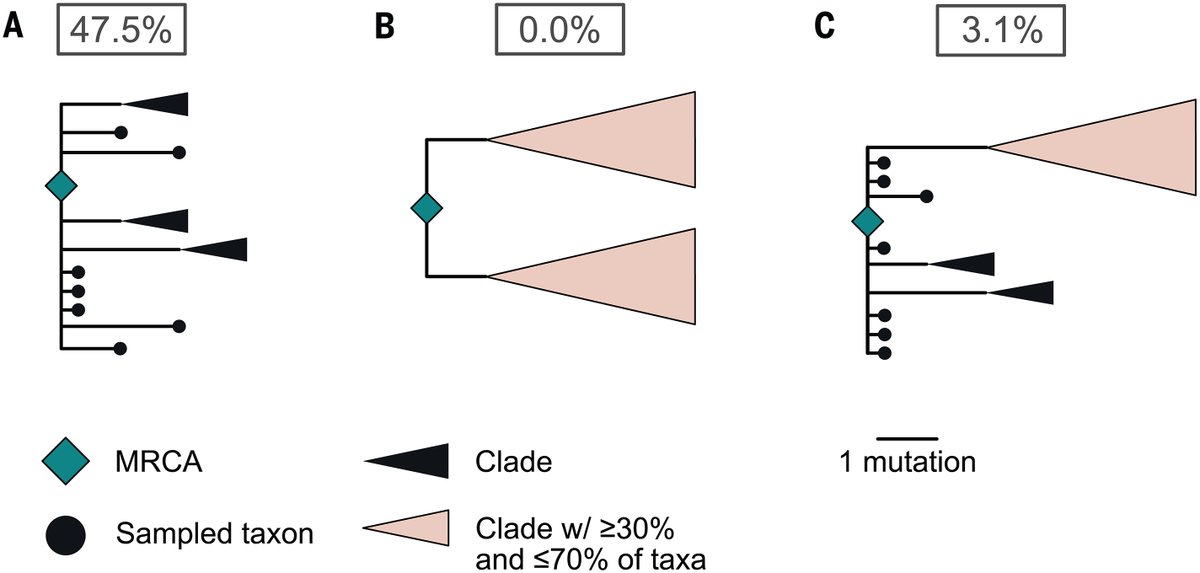
 Unfortunately, the likelihoods are distorted by errors and noise. We can correct these errors and resample to suppress noise (). This changes the picture slightly, but it still isn't useful. The likelihood on the left isn't really what we're interested in. github.com/nizzaneela/mul…
Unfortunately, the likelihoods are distorted by errors and noise. We can correct these errors and resample to suppress noise (). This changes the picture slightly, but it still isn't useful. The likelihood on the left isn't really what we're interested in. github.com/nizzaneela/mul…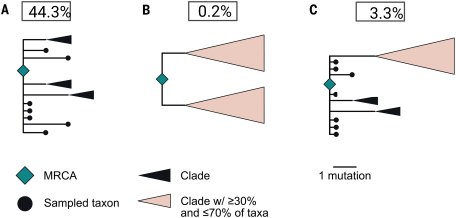
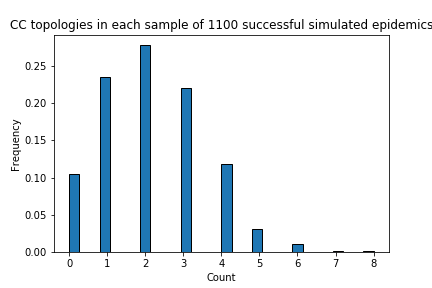
https://twitter.com/MichaelWorobey/status/1759782755431526608I'm serious. For the primary analysis, I get a 1/10 likelihood from 1000 complete resamples of the final stochastic phase (mutations generated by "stableCoalescence_cladeAnalysis.py") using the 1100 time trees from the Zenodo data. I'm worried the other four analyses...

https://twitter.com/gdemaneuf/status/1717131822277210134A lot of people were skeptical of the modelling behind Pekar '22. I was particularly worried about the way epidemics were simulated over simple networks that lacked realistic structures and instead had a scale-free degree distribution.
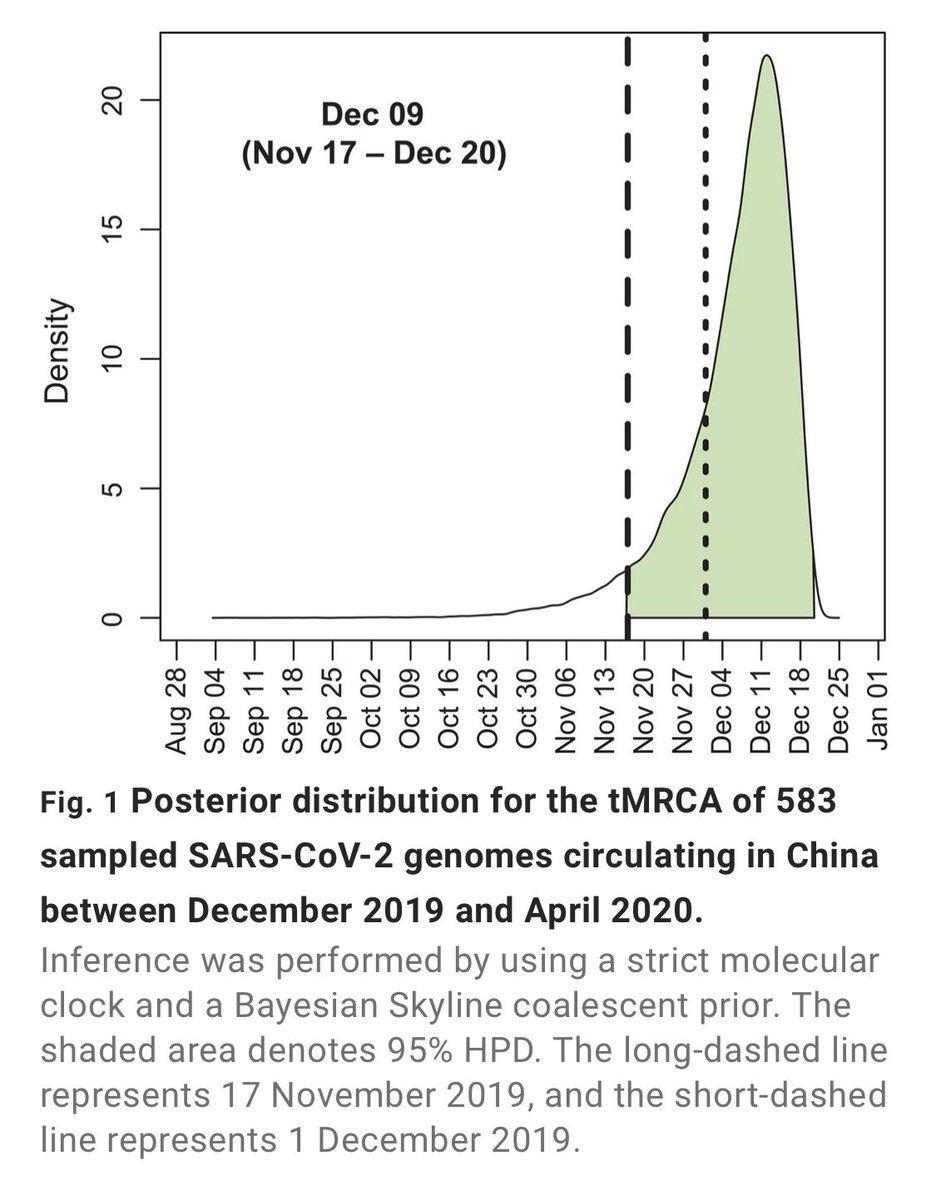
https://x.com/nizzaneela/status/1501599506139340811?s=20