
Director of bioinformatics at AstraZeneca. YouTube at chatomics. On my way to helping 1 million people learn bioinformatics. Also talks about leadership.
How to get URL link on X (Twitter) App


 1/ Why gene lengths matter: You need them for normalizing ChIP-seq signals in gene bodies (like H3K36me3). For RNA-seq, exon lengths are used to calculate RPKM or TPM.
1/ Why gene lengths matter: You need them for normalizing ChIP-seq signals in gene bodies (like H3K36me3). For RNA-seq, exon lengths are used to calculate RPKM or TPM.
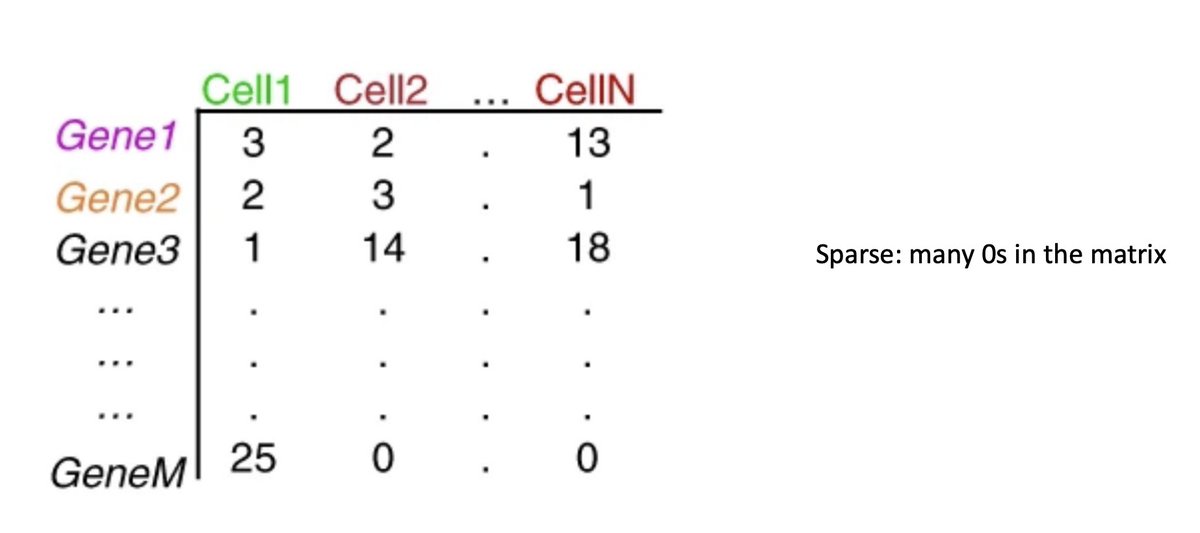
 2/
2/
 1/
1/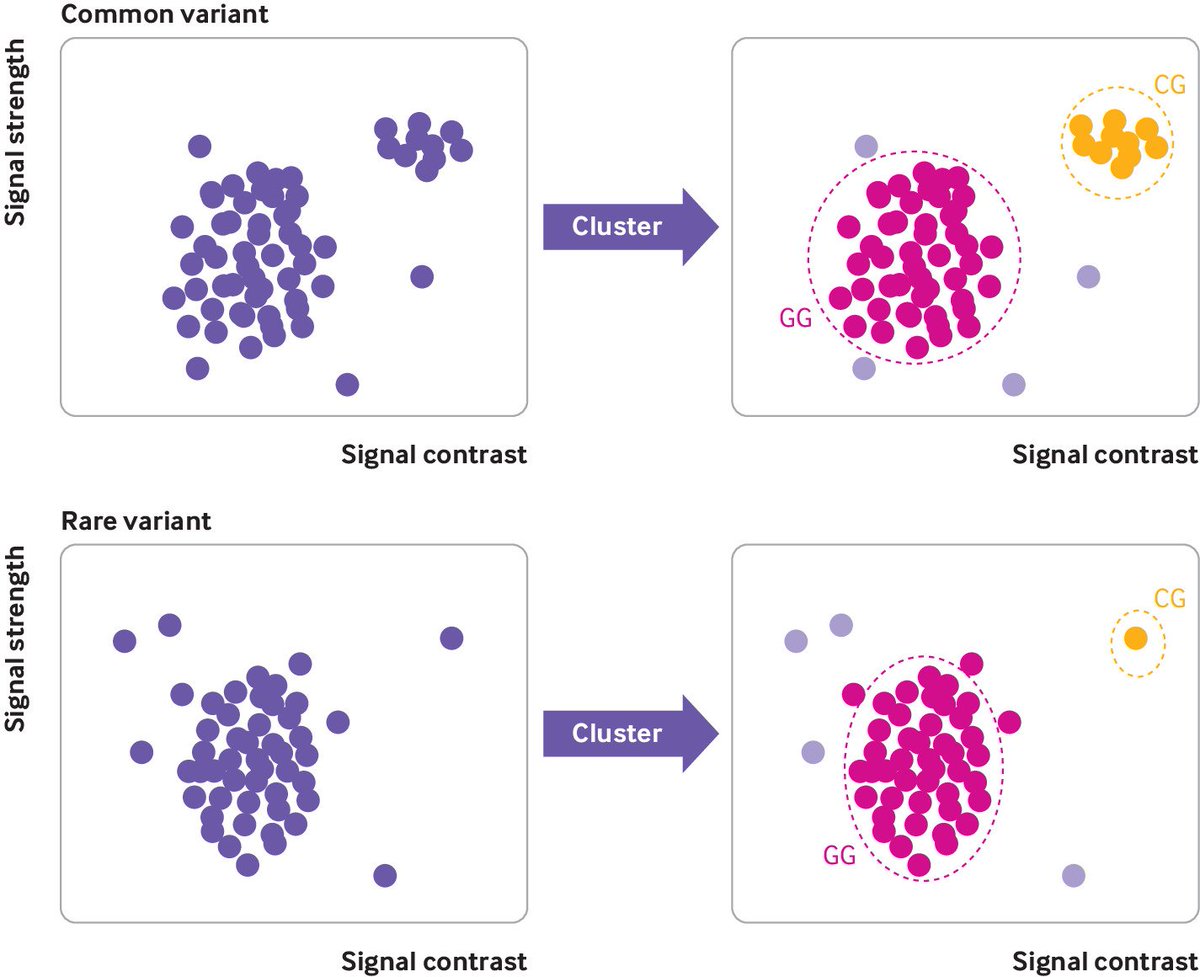
 2/
2/
 3. Modern Statistics for Modern Biology with
3. Modern Statistics for Modern Biology with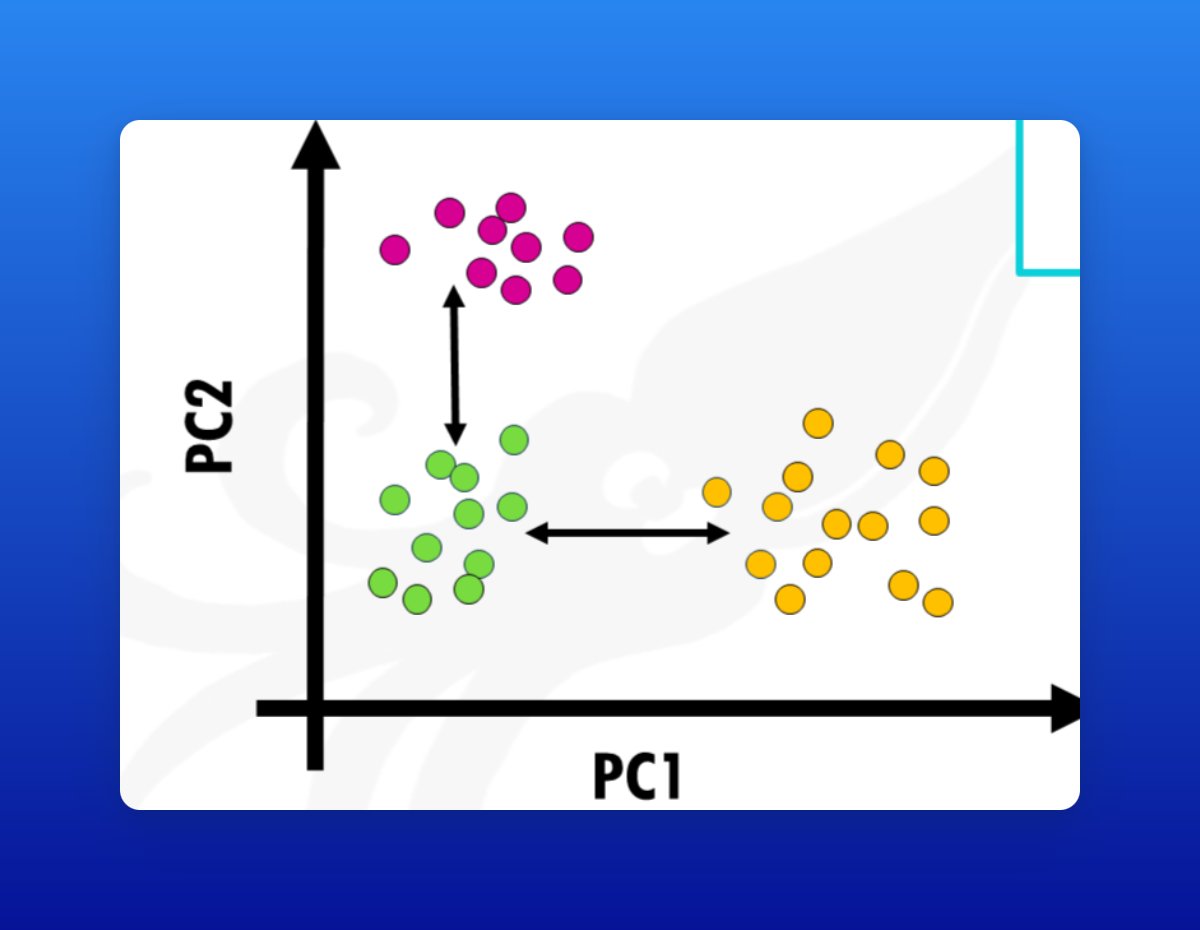
 2/ What is PCA?
2/ What is PCA?
 2/
2/
 2/
2/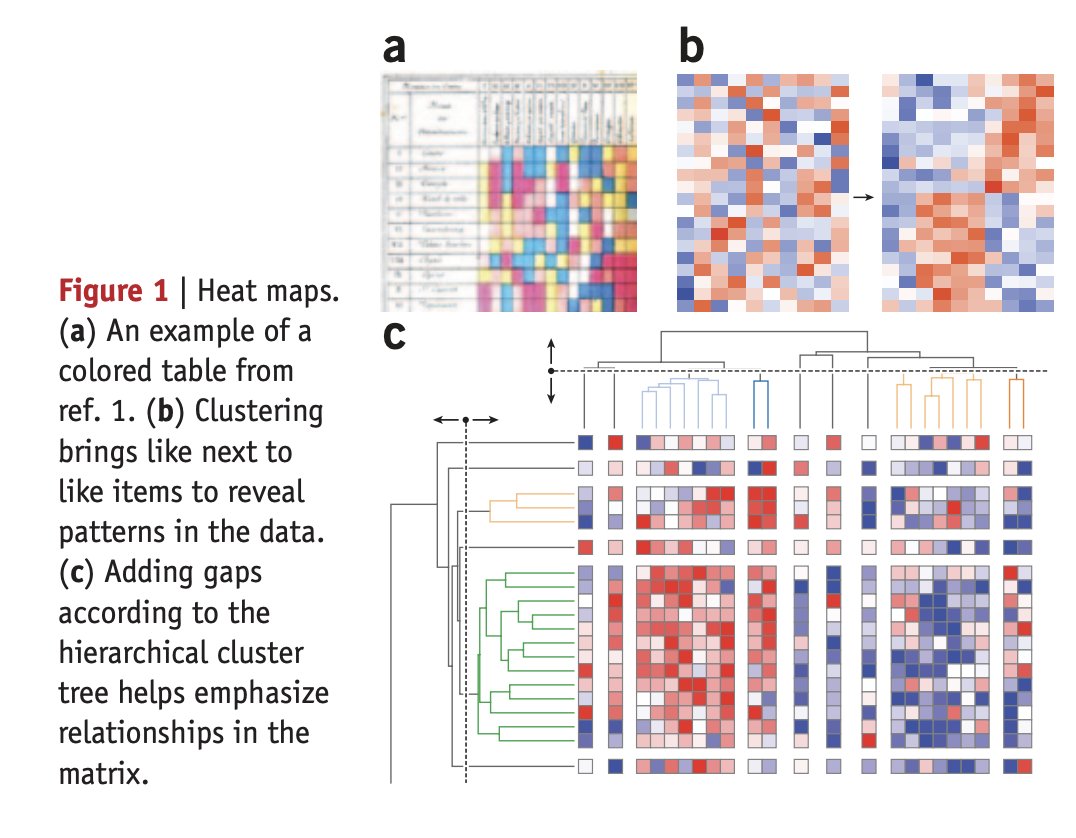
 1/ Mapping quantitative data to color nature.com/articles/nmeth…
1/ Mapping quantitative data to color nature.com/articles/nmeth…

 2/
2/
 1/ Text book: Molecular Biology of the Cell, amazon.com/Molecular-Biol…
1/ Text book: Molecular Biology of the Cell, amazon.com/Molecular-Biol…

 2/
2/
 2/
2/
 2/ There are 4 levels of bioinformatics skills, from manual work to full automation. The higher you go, the faster and more efficient you become.
2/ There are 4 levels of bioinformatics skills, from manual work to full automation. The higher you go, the faster and more efficient you become.