Insight into the spatial behavior of epidemics from @ohallats and colleagues. An initial outbreak will grow in its highly connected environment and subsequently seed outbreaks in other locations. These other outbreaks resemble a transposed copy of the first. 1/4 

@ohallats #nCoV2019 in Wuhan went from an index case in ~Nov 2019 to several thousand cases by mid-Jan 2020, thus going from initial seeding event to widespread local transmission in the span of ~10 weeks. 2/4 
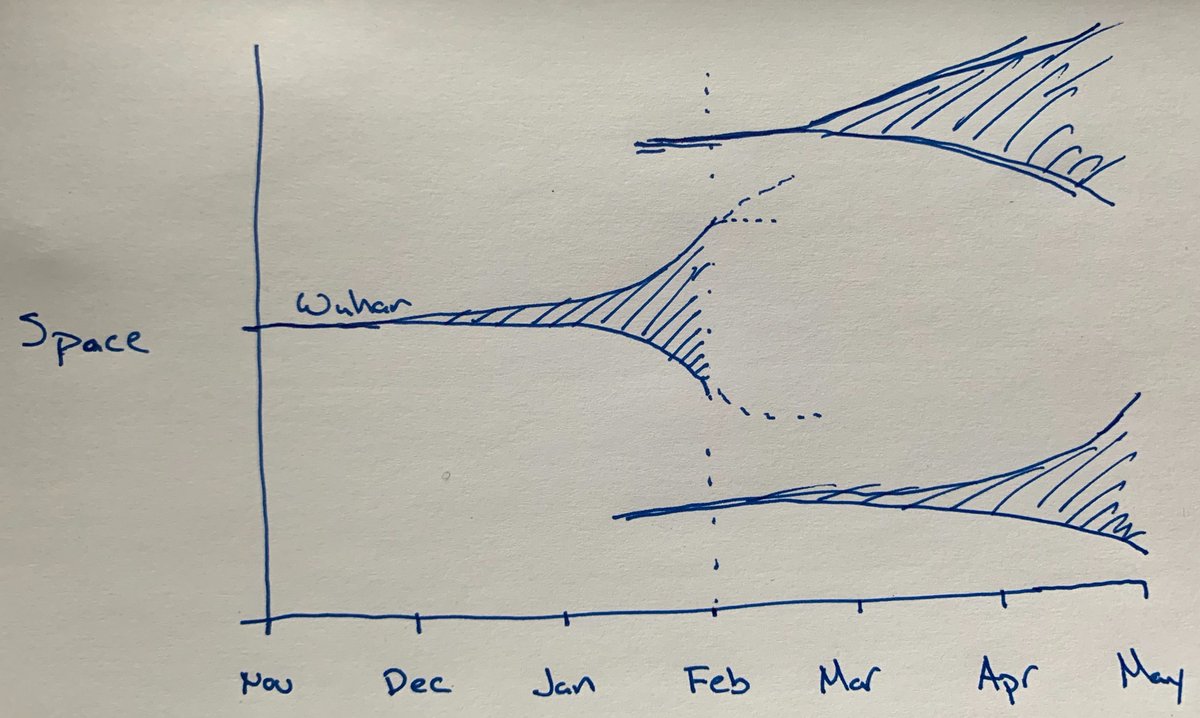
@ohallats We believe that international seeding events started to occur in mid-Jan. Thus we have a critical ~10 weeks from then to late-March to contain these nascent outbreaks before they become sizable. 3/4
@ohallats References for @ohallats's work: pnas.org/content/111/46…, journals.plos.org/plosgenetics/a… 4/4
• • •
Missing some Tweet in this thread? You can try to
force a refresh