Can I use FACS staining protocol? Should #CITEseq Abs be used at saturating conc? Can I reduce staining volume? Does cell# at staining matter? How can I minimize Background?
Tweetorial of biorxiv.org/content/10.110…
@SBKoralov @skinUCPH @NYGCtech
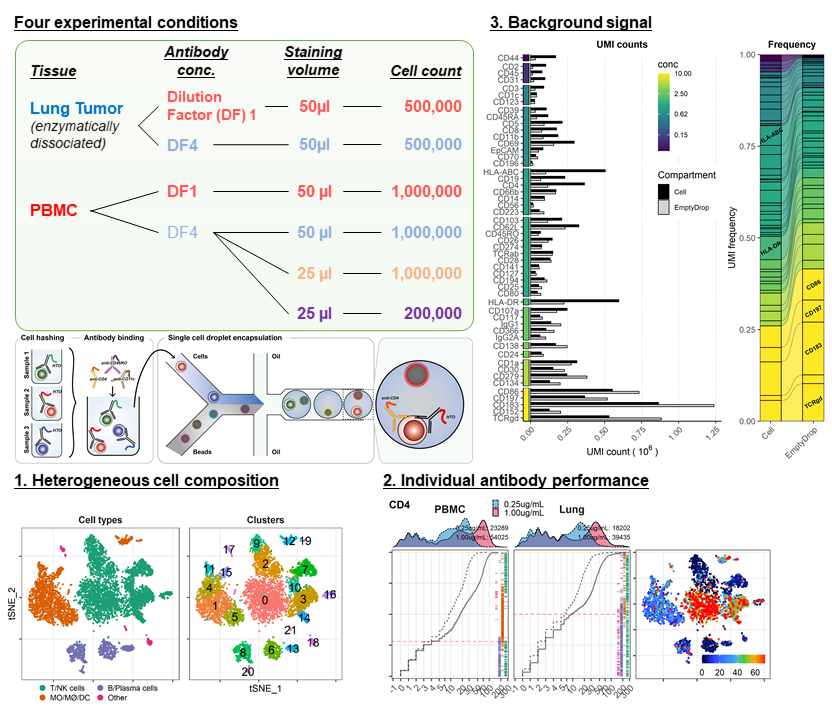
biorxiv.org/content/10.110…

Data from the study available at FigShare: doi.org/10.6084/m9.fig…
All code used for analysis and to generate the figures: github.com/Terkild/CITE-s…
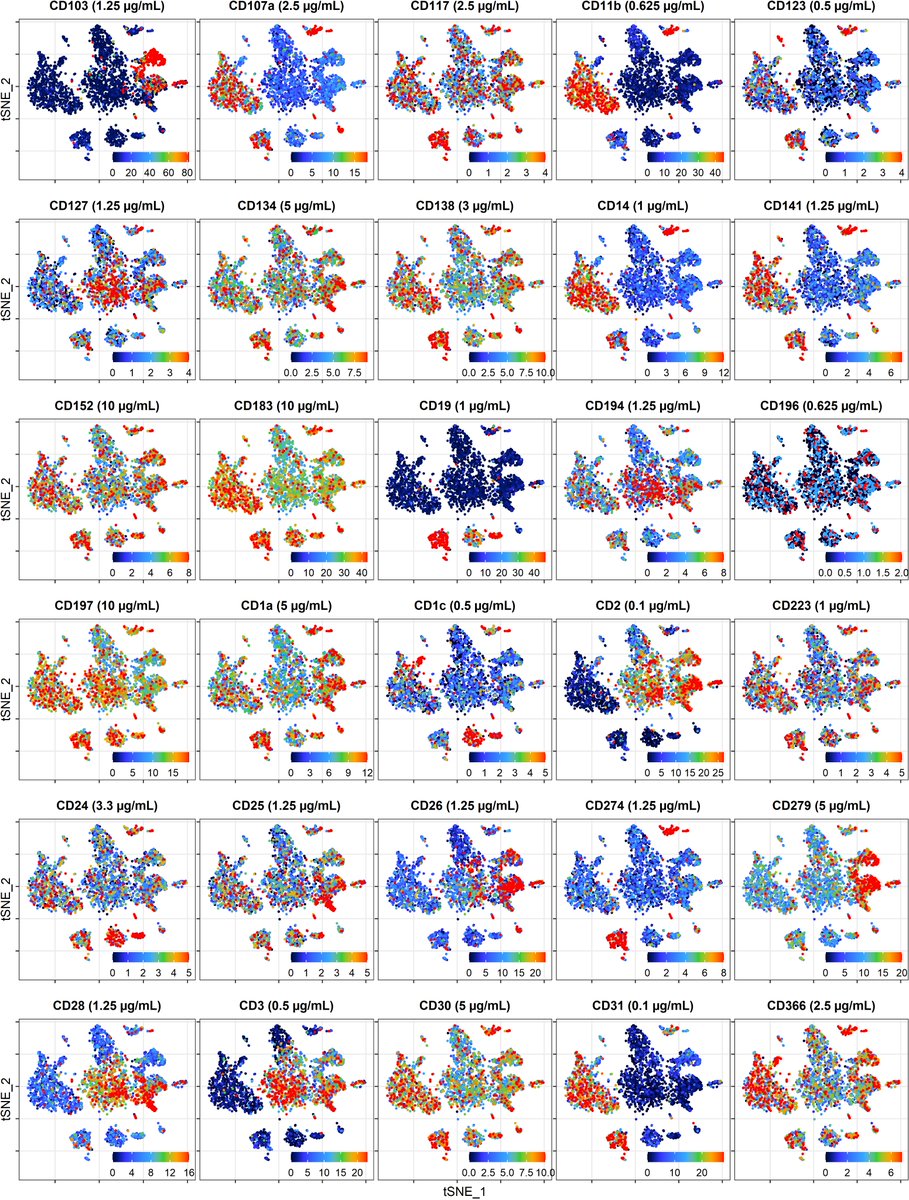
Great collaborative effort between @SBKoralov #KoralovLab, @skinUCPH, @NYGCtech (@emimitou @psmibert) & @ThalesPapaG with (hopefully) more to come!






