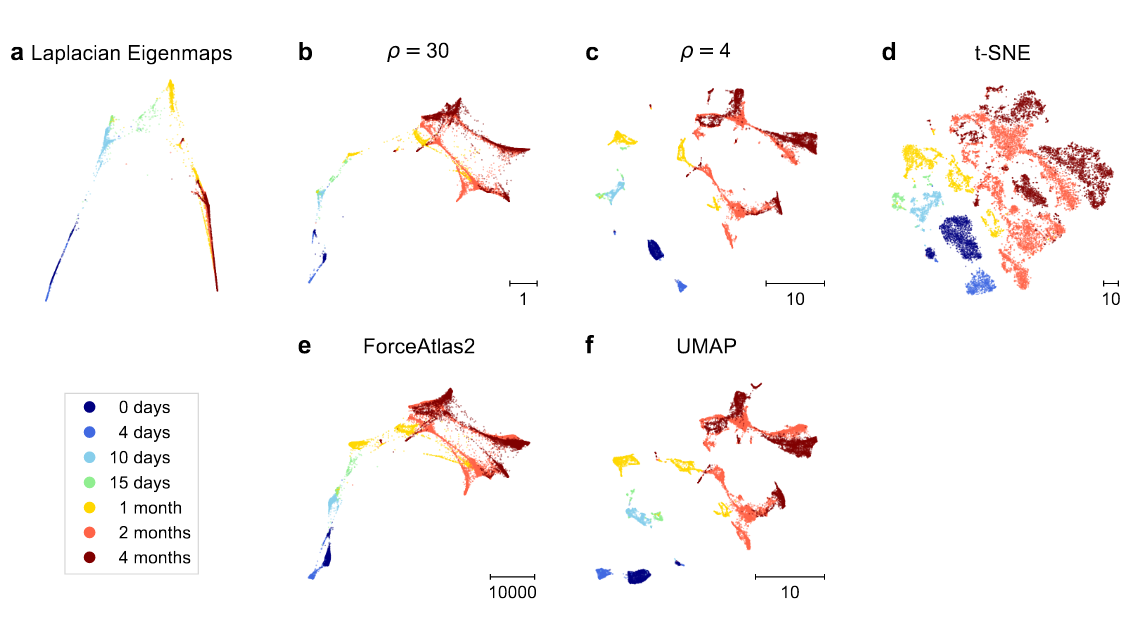
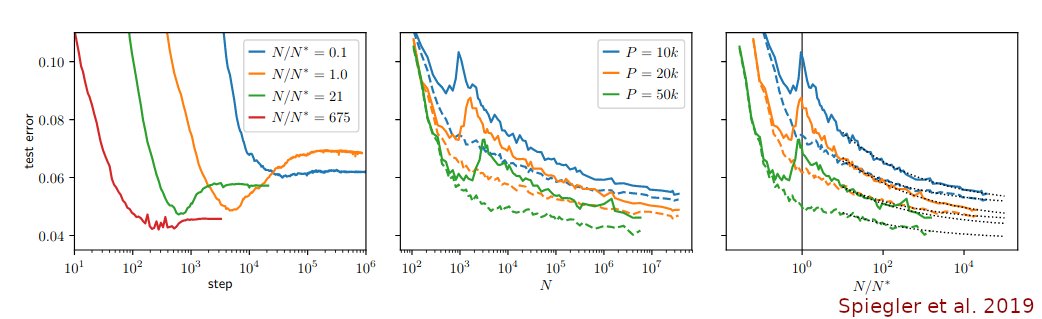
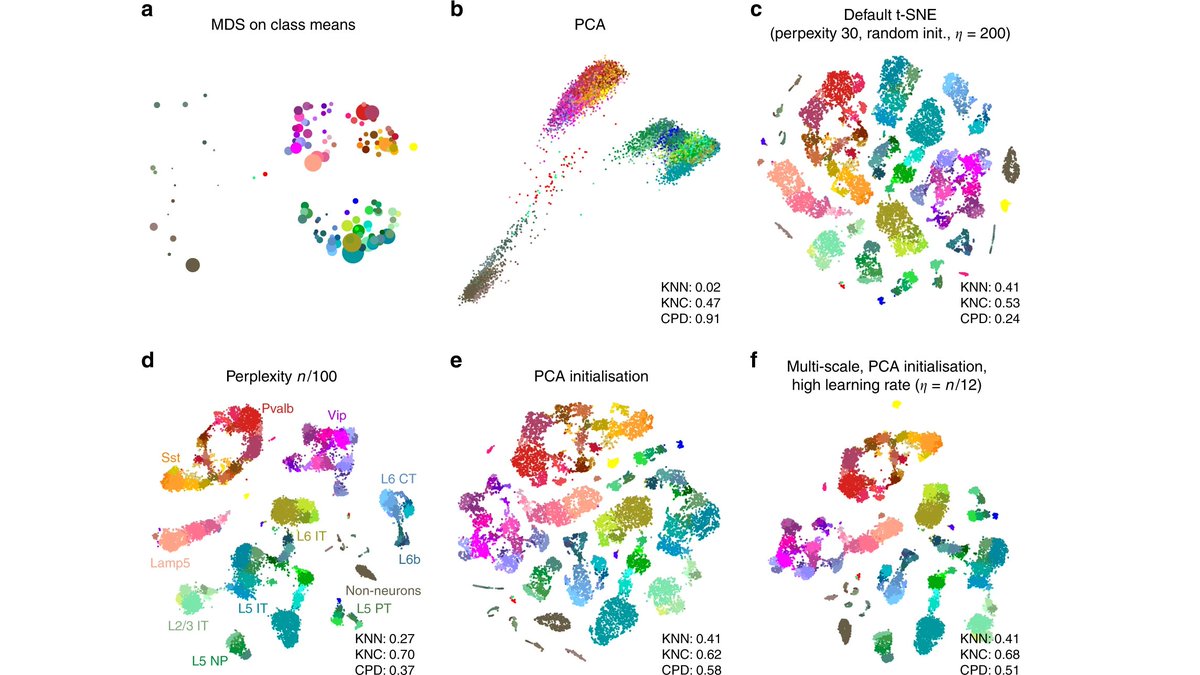
We also show that UMAP has higher attraction due to negative sampling, and not due to its loss. 🤯 Plus we demystify FA2.
With @jnboehm and @CellTypist.
arxiv.org/abs/2007.08902 [1/n]
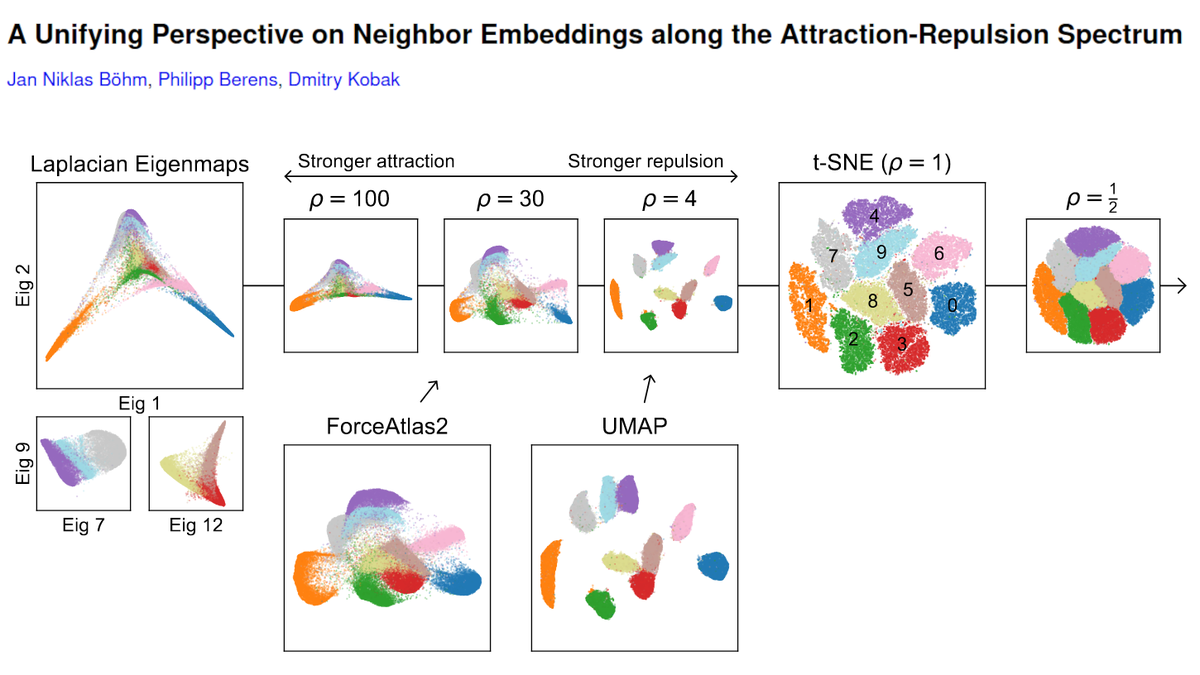
People like to use it for developmental single-cell RNA-seq data because it tends to preserve developmental trajectories better than t-SNE/UMAP. See e.g. papers by @KleinLabHMS. We show that FA2 on the kNN graph looks similar to t-SNE with ρ≈30. [7/n]