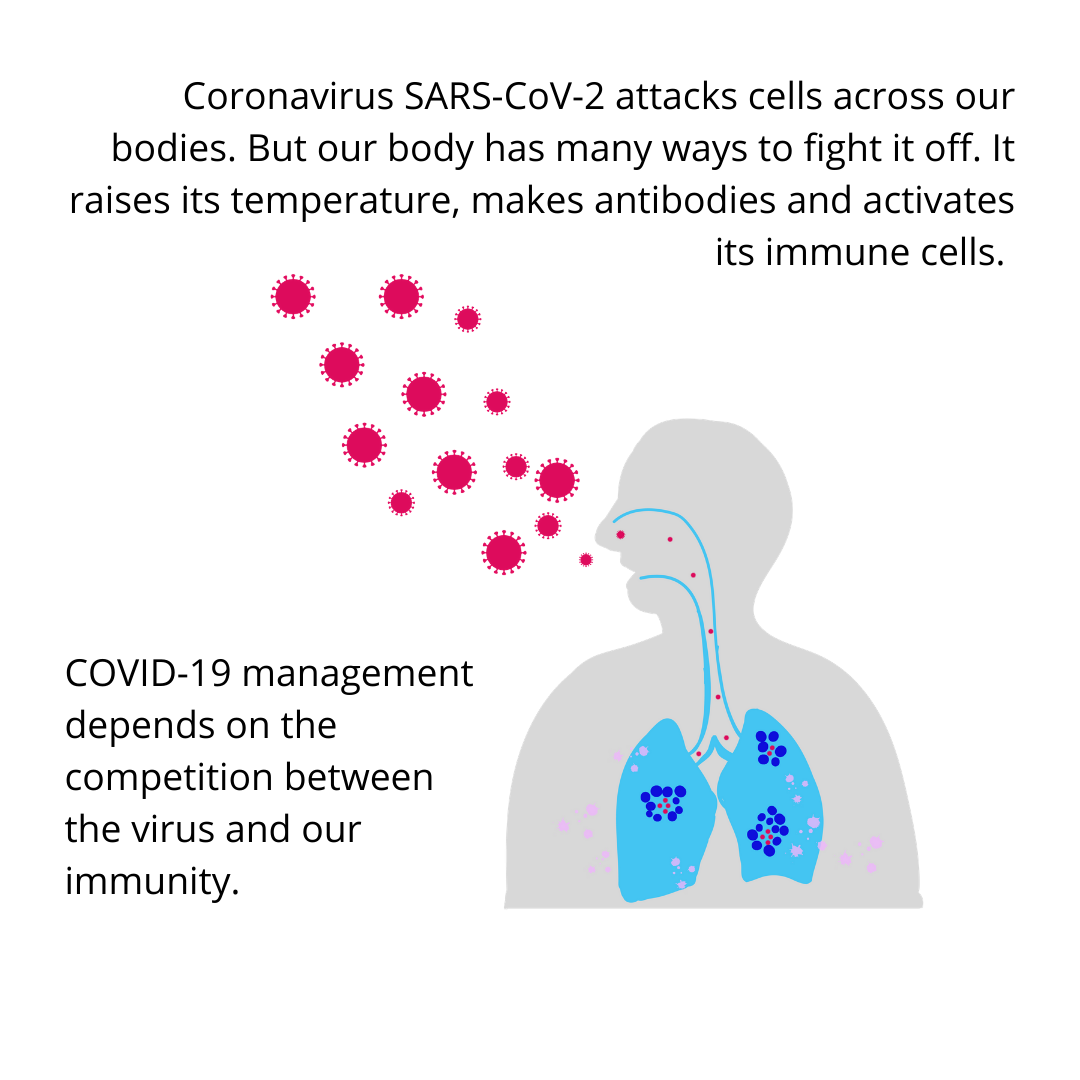
With @csiriict we did a study from 10 Sewage Treatment Plants (STP) and a gated community in Hyderabad to look for SARS-CoV-2 RNA. These STPs get 40% of sewage in the city. We estimated viral load in those samples using RT-PCR.
ccmb.res.in/presscovrg/Was…
@CSIR_IND
1/n
pubmed.ncbi.nlm.nih.gov/26246784/
Sewage treatment plants that get samples from a huge part of a city become good candidates for checking for the viral RNA from many people together.
2/n
sciencedirect.com/science/articl…
3/n
Method 1: (Ahmed et al., 2020)
No of infected individuals=((RNA copies/L water)*(L water/day)) / ((g feces/day)*(RNA copies/g feces) )
Feces excreted/person/day on an avg = 128g
One positive person sheds an avg of 10^7 RNA copies/g of feces
4/n
No of infected individuals=No of RNA copies per litre of waste water/No of RNA copies per person to total sewage water
Avg RNA copies excreted per person = 1.2×10^9
5/n
The study is submitted to @medrxivpreprint
Fin