Our paper on Compositional Tensor Factorization (CTF) of microbial dynamics is now published in @NatureBiotech! nature.com/articles/s4158…
It might change how you analyze longitudinal microbiome data. Thread below:
It might change how you analyze longitudinal microbiome data. Thread below:
This work was a great collaboration and fun with
@cameronmartino and @lisa55asil, George Armstrong, @mcdonadt, @yoshikivb, @jamietmorton, Lingjing Jiang, Maria Gloria Dominguez-Bello,@CMI_Austin, @halperineran, @KnightLabNews. Thank you!!
@cameronmartino and @lisa55asil, George Armstrong, @mcdonadt, @yoshikivb, @jamietmorton, Lingjing Jiang, Maria Gloria Dominguez-Bello,@CMI_Austin, @halperineran, @KnightLabNews. Thank you!!
In a cross-sectional study, you run a PCoA and look at the top PCs. But with temporal data, applying PCoA separately on each time point may mask important information that is carried over time.
For your longitudinal data analysis you should use CTF!
For your longitudinal data analysis you should use CTF!
CTF is an unsupervised dimensionality reduction method tailored for repeated measures.It decomposes your temporal or spatial microbiome data into loading vectors corresponding to subjects, microbial features, and time/space - while accounting for its compositionality. 

In simulations, generated based on real longitudinal data distributions, CTF showed higher accuracy than existing beta-diversity metrics including Jaccard, Bray–Curtis, Aitchison, unweighted UniFrac and weighted UniFrac. 

With real data, tracking infant gut development over time, CTF was *tenfold* more accurate than other beta-diversity metrics at discriminating vaginally from cesarean born infants based on their microbiomes. 

The cool part is that CTF identified the same taxa as being associated with birth mode across independent studies, suggesting a birth mode signature across infants. These datasets were collected and processed using different protocols and by different laboratories! 
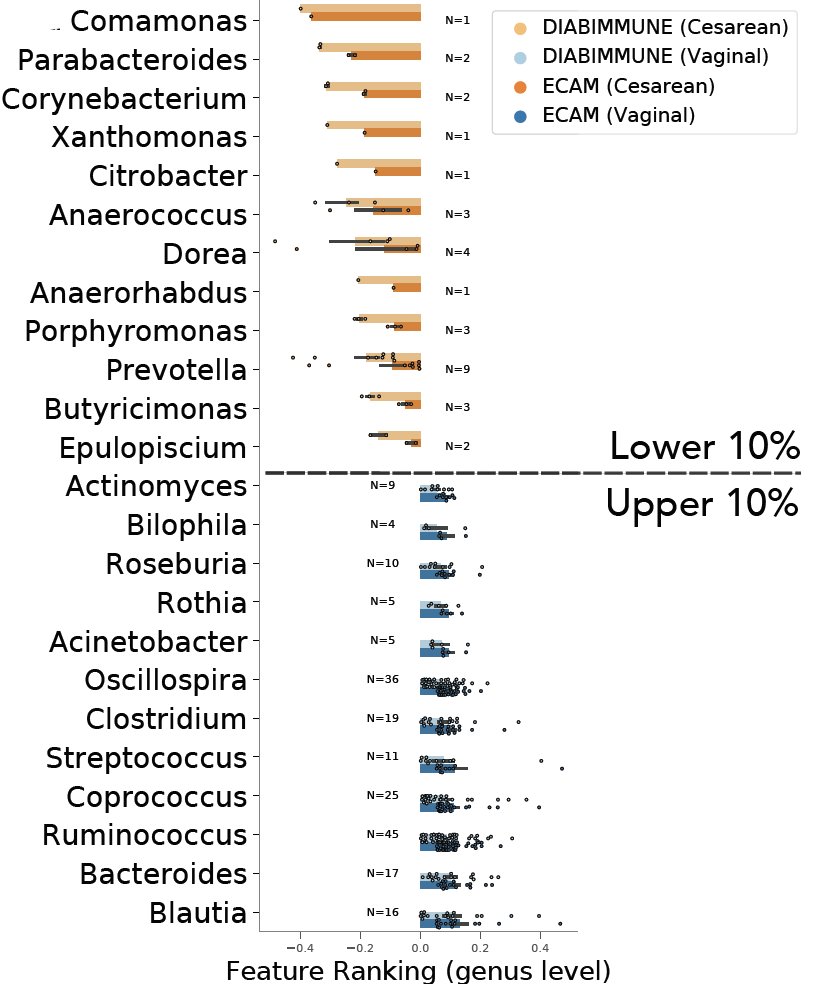
The robustness of this microbial signature across multiple datasets shows that CTF can be used to identify important players in the microbiome that are associated with host phenotypes. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh






