Excited to share our work using single-cell RNA sequencing to profile the lung parenchyma in advanced COPD. Follow 🧵for highlights and new #COPD insights medrxiv.org/content/10.110…
Manuscript is co-authored with @jmcdonou, w/ leadership from @KaminskiMed @YalePCCSM and Ivan Rosas @bcmhouston and important contributions from Taylor Adams, Neeha Kothapalli @Yale @ptimshel @jonas_schupp @caroymalo @JLGomezMD @SergioPoli @mauchioccioli & others not on twitter
We identified major cell types from 15 control donor lungs and 17 explanted lungs from COPD patients requiring transplant and builds on the previous study by Adams et al advances.sciencemag.org/content/6/28/e… 

We identified two types of AT2 cells with similarities to AEP and non-AEP described by @DrWillZacharias, Mark Krasnow, and others although mostly separated by the degree of surfactant production. So we use the modular terms AT2A(~AEPs, low surfactant) and AT2B (high surfactant) 
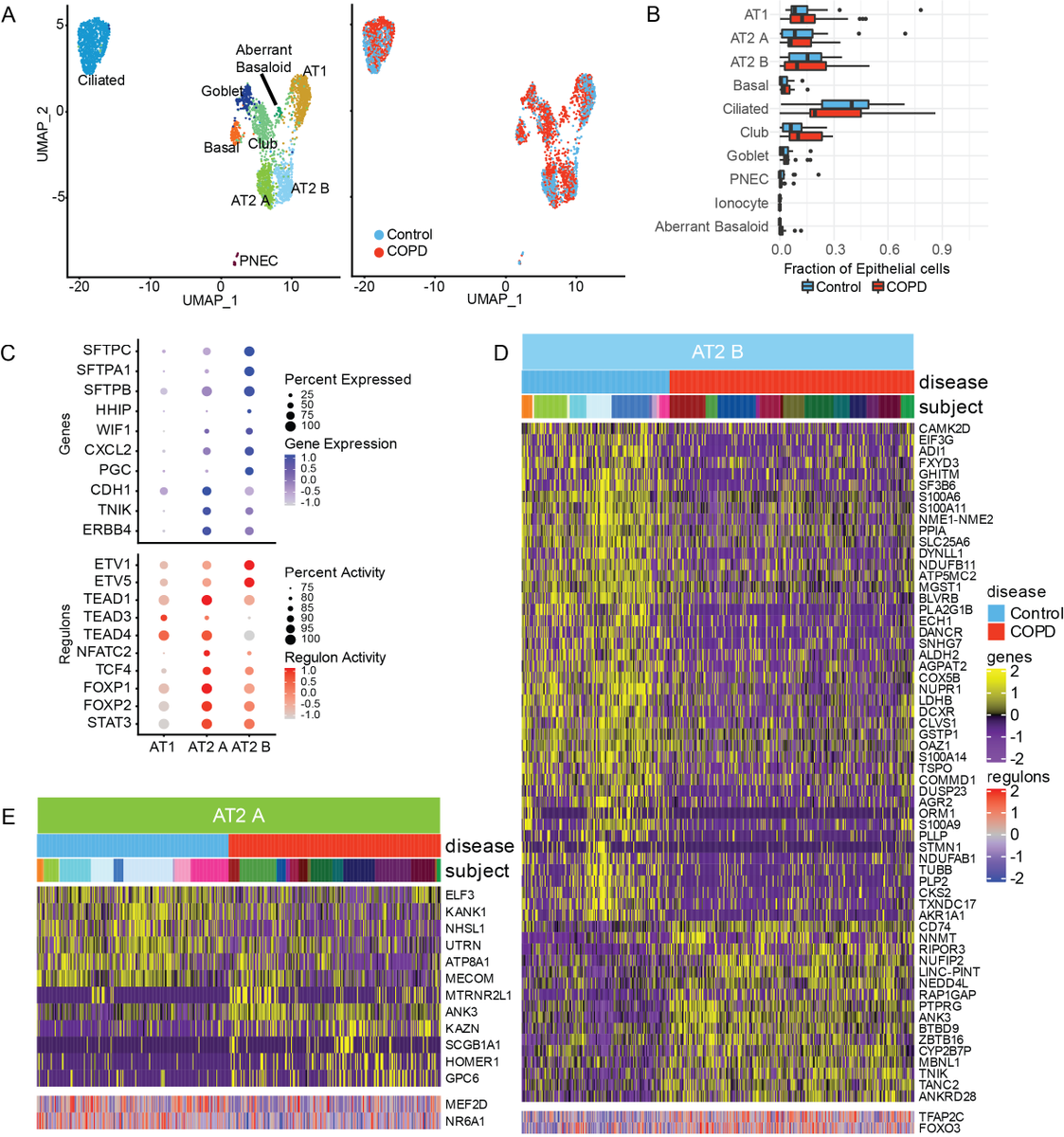
Interestingly, with advanced COPD, the AT2Bs have decreased expression of cellular stress response genes while AT2As have decreased expression of genes related to cellular differentiation. These divergent changes may underlie their role in COPD pathogenesis.
We also found the lung endothelium in COPD is on🔥 There were expression changes across multiple endothelial subtypes consistent with endothelial activation, with 👆xexpression of genes related to inflam, and 👇expression of angiogenesis and endothelial homeostasis genes. 

We also identified changes in macrophages population composition in advanced COPD, particularly an increase in metallothionine expressing macrophages. 

We used CELLECT, by @ptimshel, which co-localizes data from GWAS studies and scRNAseq. We integrated the scRNAseq data with UKBiobank GWAS studies by
@mikecho95 and made the interesting observation that non-hematopoietic cells mediate much of COPD heritability.
@mikecho95 and made the interesting observation that non-hematopoietic cells mediate much of COPD heritability.

Finally, we applied the lung connectome, developed by @msbr89, to generate network-level maps of alveolar signaling in control and COPD. We found changes in multiple canonical pathways, with the most striking change occurring in CXCL-motif chemokine signaling 

Thank you to the incredible environment at
@YalePCCSM @YaleIMed and @YaleMed as well as @nih_nhlbi 3 Lakes Partnerships, and @YaleGeriatrics for providing generous support, and @KaminskiMed for the opportunity to work on this project.
@YalePCCSM @YaleIMed and @YaleMed as well as @nih_nhlbi 3 Lakes Partnerships, and @YaleGeriatrics for providing generous support, and @KaminskiMed for the opportunity to work on this project.
If you want to explore the scRNAseq data and identify changes with advanced COPD, you can go to copdcellatlas.com Great for members of @ATS_RCMB @atscommunity @ATS_AII @COPDFoundation
• • •
Missing some Tweet in this thread? You can try to
force a refresh






