
Very excited to share our updated preprint on pooled testing for SARS-CoV-2 surveillance. This has been a fantastic modeling and lab collaboration with @BrianCleary, @michaelmina_lab and Aviv Regev, and it’s all about our favorite topic: viral loads. 1/16
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
Highlights:
-PCR sensitivity and efficiency are linked to epidemic dynamics and viral kinetics
-Prevalence estimation without testing individual samples using a few dozen tests
-Simple (by hand) strategies optimized for resource-constrained settings
Full story below. 2/16
-PCR sensitivity and efficiency are linked to epidemic dynamics and viral kinetics
-Prevalence estimation without testing individual samples using a few dozen tests
-Simple (by hand) strategies optimized for resource-constrained settings
Full story below. 2/16
We (the world) still need more testing. The number of test kits is still limited in a lot of places, meaning that we are missing a lot of infections, not testing regularly, and are flying blind wrt population prevalence. Pooling has been discussed as part of the solution. 3/16
Pooling is simple – mix samples together and test the pool. If -ve, assume all constituent samples were negative and stop. If +ve, re-test the original samples to find the positives. When most pools test -ve, we use fewer tests than needed to test each individual sample. 4/16 
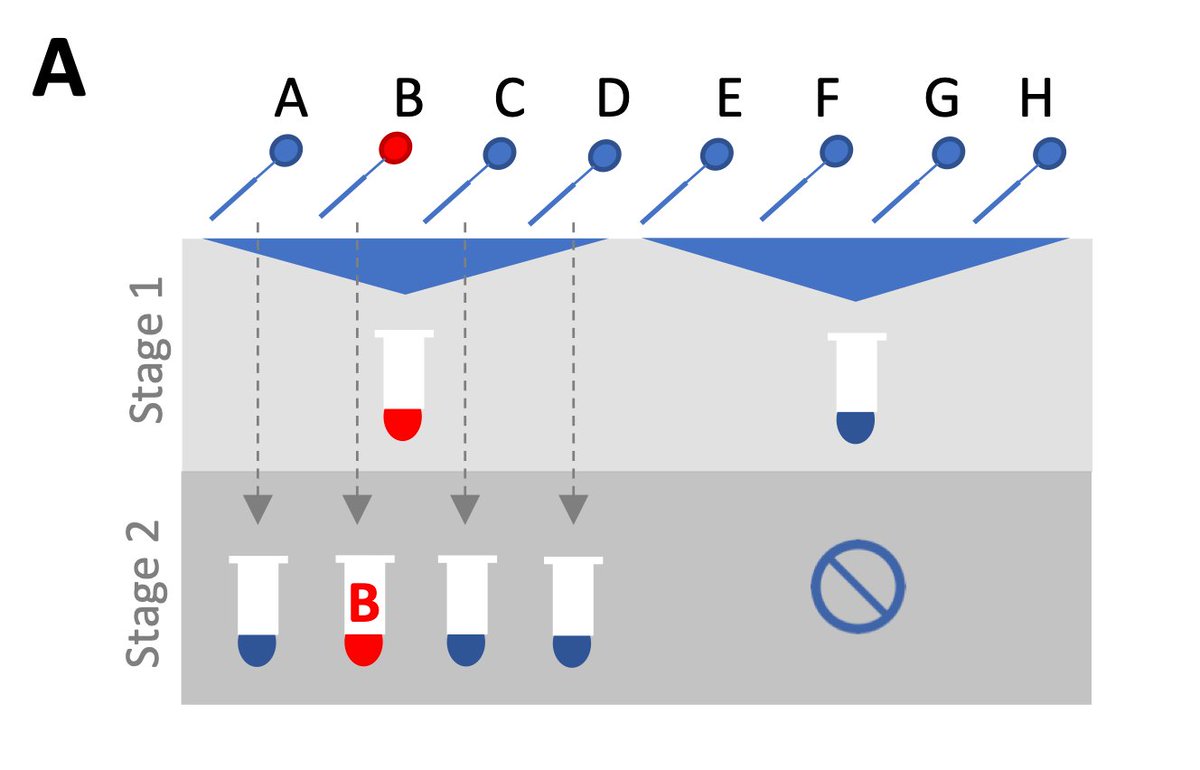
But there are a few key concerns with pooling: 1) we might miss low-viral load samples through dilution effects, 2) efficiency changes depending on prevalence, and 3) complicated pooling strategies are more logistical hassle than they’re worth, particularly without robots. 5/16
A lot of papers/preprints have investigated these issues and propose clever pooling algorithms. But there is a missing component: viral loads vary over the course of infection, between individuals and over the epidemic*, described brilliantly here: nytimes.com/interactive/20…
6/16
6/16
* This is a really interesting point, but a story for a different thread.
To model this, we fit a random-effects viral kinetics model to time-series viral loads. We then used an SEIR model to simulate loads of infections and individual-level viral load curves. This gave us a synthetic population of viral loads to test pooling strategies. 7/16 
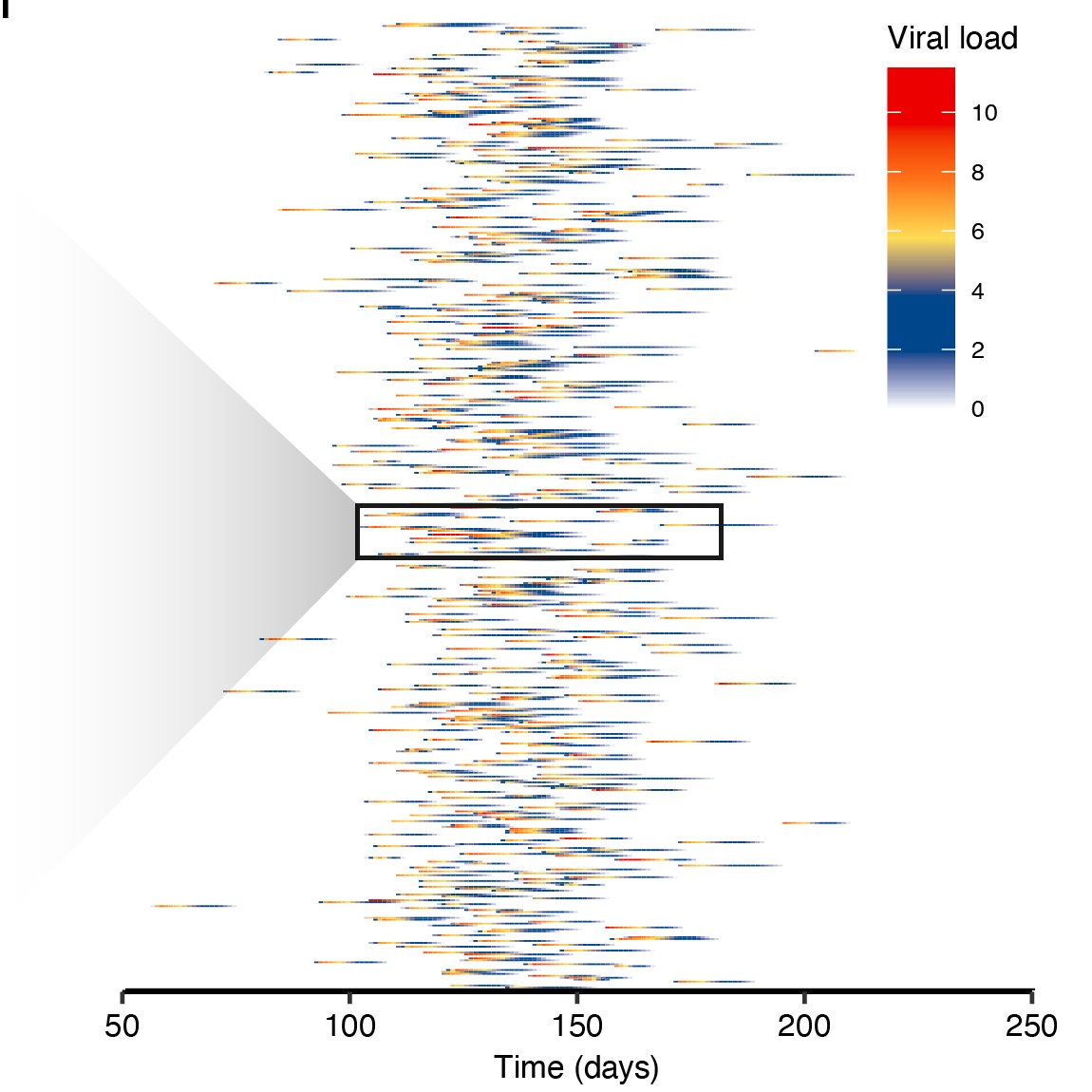
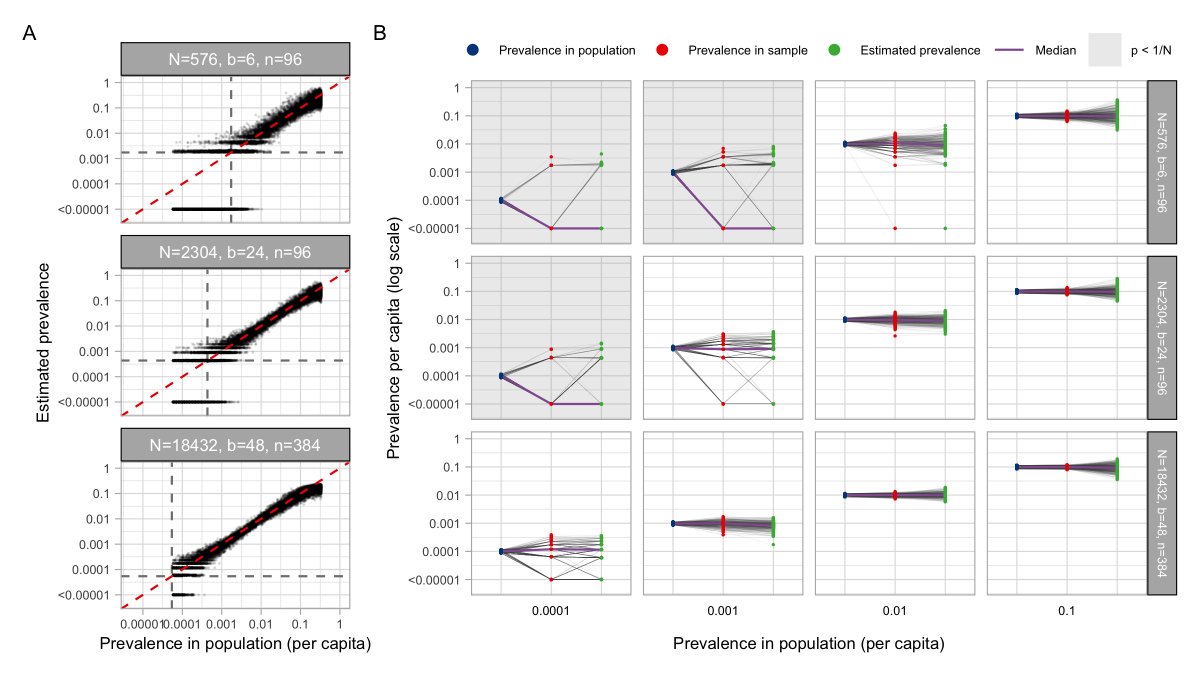
For prevalence estimation, we used a statistical method developed for HIV. You write down a likelihood for the viral load observed in a pool *given* prevalence, and use this method to get a maximum likelihood prevalence estimate based on the viral loads of your tested pools. 8/16
With enough samples (if prevalence is low, need more samples to capture >0 positives), the method gives accurate estimates of prevalence using <=48 tests. We checked this in the lab – we were able to estimate 1% prevalence amongst ~2000 samples using only 48 tests! 9/16 
For individual testing, we identified pooling strategies that were a) simple to carry out, b) would remain efficient even if prevalence changed due to epidemic dynamics, and c) maximized the number of positive samples identified when testing capacity is limited. 10/16 
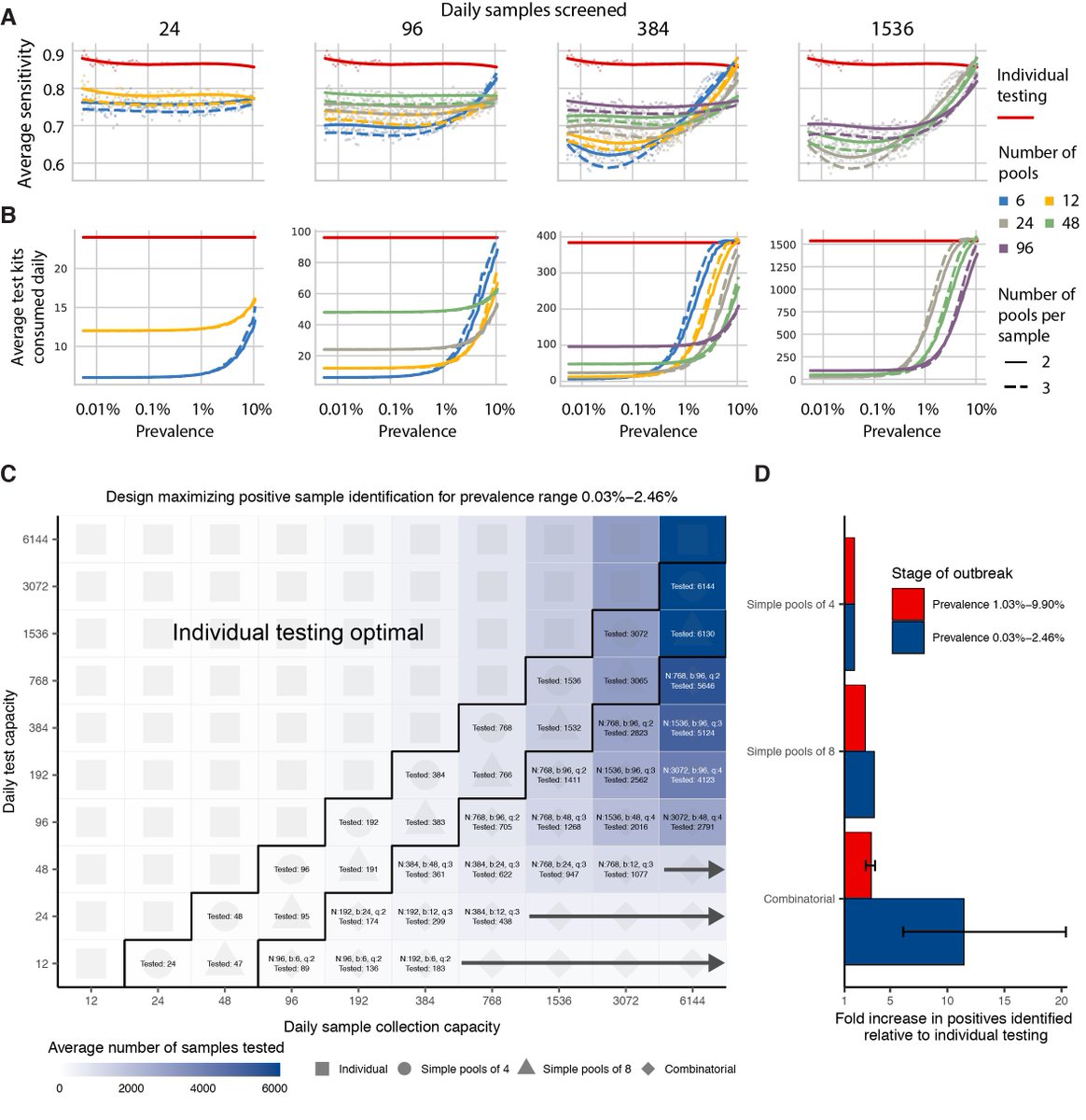
We did exactly the same thing comparing the growth and decline phases of the epidemic (sensitivity is generally lower during epidemic decline, because more samples have low viral loads), and also using simulations based on sputum samples. 11/16
Because we knew the infection status and true viral loads of our entire population, we could look at sensitivity depending on when during an infection someone is sampled. Unsurprisingly, most false negatives come from samples in the tail end of their infection. 12/16
Finally, we tested our theoretical predictions in the lab using discarded nasopharyngeal swabs. Pooling samples of varying viral loads gave results consistent with expected dilution effects, and we showed that simple pooling led to efficiency gains in line with expectation. 13/16 
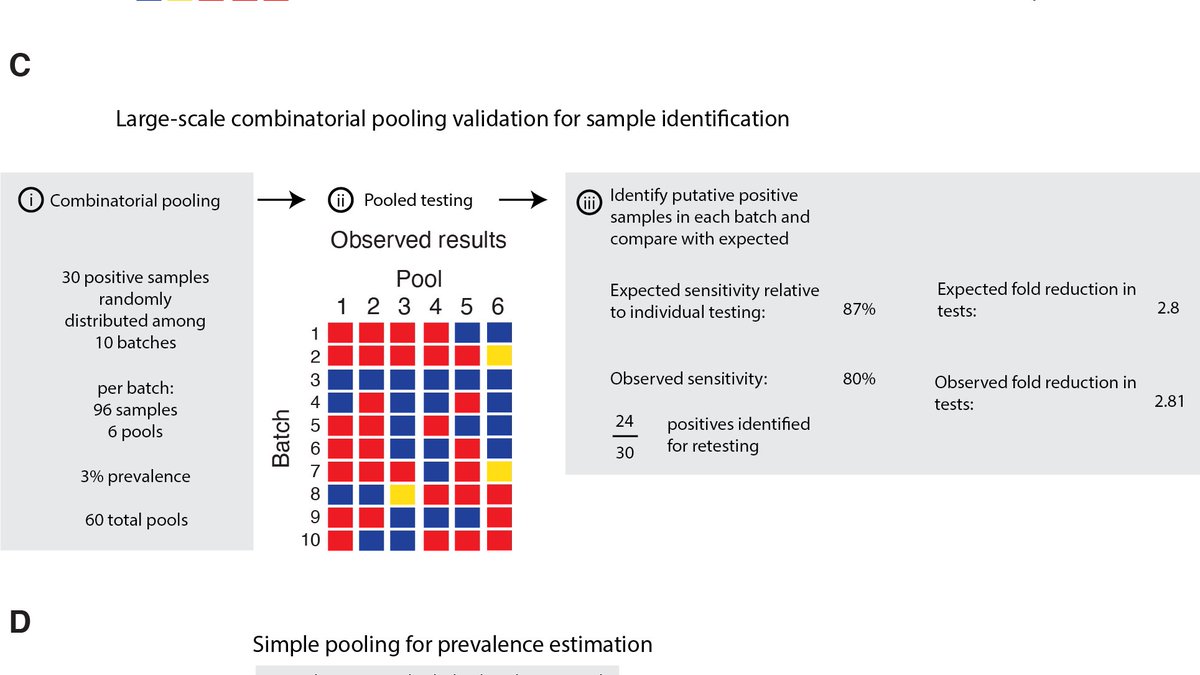
Take home: simple protocols that can stay unchanged for weeks drastically increase the number of +ves identified. Loss of sensitivity plays out as expected with dilution effects – most missed samples are from low-viral load individuals, mostly at the end of their infection. 14/16
Whether to pool or not depends on your question and setting. If test kits are limited, you will likely identify more +ves overall by pooling, and the small sensitivity loss may therefore be tolerable. Settings aside a few tests for prevalence estimation is also a good idea. 15/16
Lower sensitivity may not be tolerable in a clinical setting, so whether to pool or not depends on how the test result will change triaging. But for surveillance and sheer throughput, simple pooling is the way! 16/16
Huge thanks to all those involved in the study: Brendan Blumentstiel, Maegen Harden, Michelle Cipicchio, Jon Bezney, Brooke Simonton, David Hong, @m_senghore, @DocKarim221 and Stacey Gabriel.
• • •
Missing some Tweet in this thread? You can try to
force a refresh


