
#ISTC20 #Sesh5 1/16 I thank the organisers for a chance to present my research here. I work on the Spoon-billed sandpiper (SBS), a charismatic species, characterised by an unusual shape of its beak. It is also critically endangered, with <200 pairs left in the wild. 

#ISTC20 #Sesh5 2/16 SBS breeds only in Chukotka (the most North-Eastern part of Eurasia). It flies to Southeast Asia for winter, stopping at key sites in China and other countries during the trip. It is a flagship and umbrella species helping to shape environmental policy. 

#ISTC20 #Sesh5 3/16 We supplemented the headstarting efforts of this species with a thorough genomic analysis. To this end, we sequenced ~10 SBS complete genomes from different locations along the breeding range. The quality of the reference genome is comparable to chicken. 

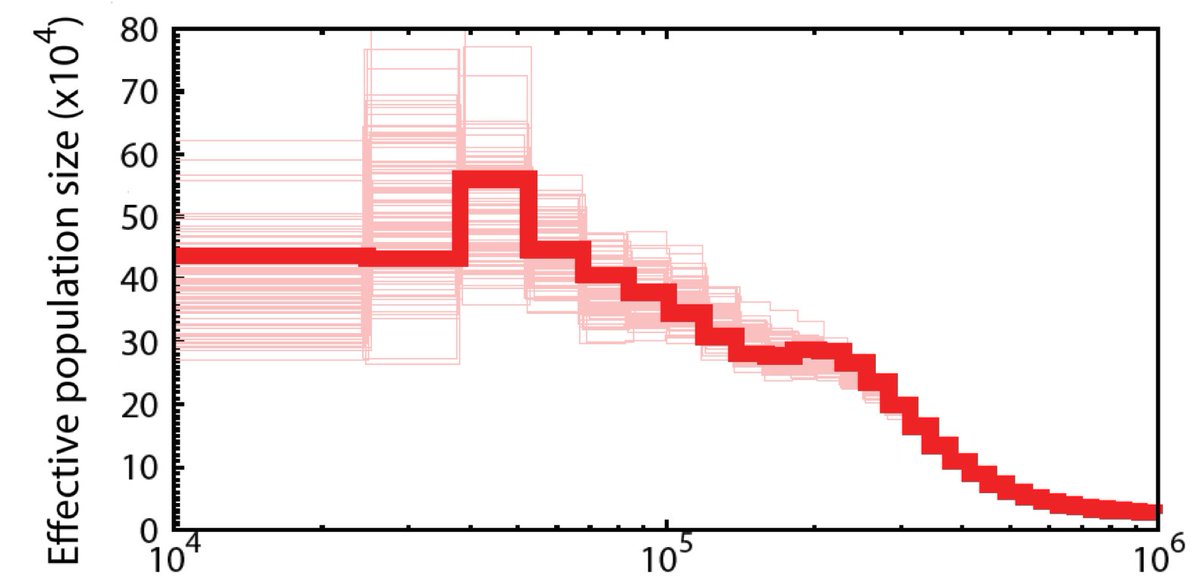
#ISTC20 #Sesh5 4/16 Using the polymorphisms found in the reference genome we can perform a PSMC analysis, which reconstructs the demographic history of the species. We found that SBS peaked at ~20 tya, and continued to decline since. 

#ISTC20 #Sesh5 5/16 20 tya is the time of the Last Glacial Maximum. At that time, we predict that a much wider area of the Beringia and Okhotsk sea had suitable habitat for SBS. We do not know about the possible change of habitat on the flyway or the wintering grounds. 

#ISTC20 #Sesh5 6/16 We see that the SBS population is homogeneous. SBS from Belyaka spit is just as different from another SBS on the Belyaka spit as it is from SBS from Meinypylgino. This suggests that the fidelity of specific pairs is counteracted by panmixis among young birds. 

#ISTC20 #Sesh5 7/16 The real power of genomics in in comparison. Thus, we sequenced >10 genomes of closely related species. We demonstrate that the Red-necked stint (RNS) is the sister species with little if any intergression between SBS and RNS (data not shown today). 

#ISTC20 #Sesh5 8/16 RNS had a different demographic history that SBS: it always was ~10 times more abundant than SBS (numbers older than 0.5 mya not reliable). Because of constant population size we can assume equilibrium of polymorphisms in RNS (this is important and powerful). 
#ISTC20 #Sesh5 9/16 RNS has always been 10 times more abundant than SBS. Yet their levels of heterozygosity (het) are way too similar. This cannot be explained if the segregating polymorphisms are purely neutral (pi would have been 10 times different then). What gives? 

#ISTC20 #Sesh5 10/16 Distribution of selection coefficients of new mutations may give similar het in two species with Ne differing 10-fold. We assume a skewed normal distribution (mean μ, variance σ, skew α) and that dominance related to selection as in the figure. 

#ISTC20 #Sesh5 11/16 Because of equilibrium in RNS, we use a simple formula to solve for het for these 4 parameters (μ σ α β). In SBS that is impossible, but now we know what sort of ranges we must consider. The RNS het solution is done in Mathematica in a few of hours. 

#ISTC20 #Sesh5 12/16 Because SBS is not in equilibrium, het and number of derived SNPs per genome (density) are different. Using an individual-based model in C++ we see what areas of parameter space where 1) RNS het 2) SBS het and 3) SBS density all matched the observed values. 



#ISTC20 #Sesh5 13/16 Here are the regions of solution space where all three parameters can be explained: RNS het, SBS het and SBS SNP density. The individual-based model for SBS takes days on a massively parallel computer to run. We are working on a wider area of parameter space. 

#ISTC20 #Sesh5 14/16 These are the distributions of selection coefficients of new mutations that fit all the data. The shape of that distribution is a fundamental question in evolutionary genetics. We could do it only by comparing SBS and RNS (with constant RNS pop size). Luck... 

#ISTC20 #Sesh5 15/16 We learned: SBS is panmictic, it has a high het, it was abundant during the last glacial maximum. Most importantly, combining luck and good comparative genomic design we answered a long-standing question in genetics. Making SBS a model pop gen species. 

#ISTC20 #Sesh5 16/16 Too many people to thank for Twitter. The key people are Mateusz Konczal @MateuszKonczal and Anastasia Lyulina @aIyuIina. The paper has 32 authors from 9 countries. Key funding and institutions in the figure. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh


