
Proud of our 1st manuscript! Cell2location: a new tool that integrates single cell + spatial transcriptomics data to map tissue atlases at scale. Also new biology in brain/lymph node. Fantastic collab with @OliverStegle, lead by @vitaliikl biorxiv.org/content/10.110…
(thread)
(thread)
Single cell + spatial RNAseq is highly scalable for cell atlassing. However, complex tissues present big challenges for computational methods: need to accurately map dozens of cell types including rare & fine subtypes + account for tech differences between sc/spatial data 

Enter cell2location. First, we derive cell type gene expression signatures from scRNAseq. Then, we decompose spatial data into cell type abundance maps using these reference signatures. We show c2l accurately maps two complex tissues: the mouse brain and human lymph node 

Under the hood, cell2location is a principled Bayesian model of spatial mRNA counts (NB distributed) that accounts for sc/spatial tech differences & similar locations of cell types. It can jointly model multiple batches & handle large datasets via GPU acceleration
In the mouse brain, we spatially map 56 neuronal/glial cell types using paired single nuclei + Visium RNAseq. We proceed to build our tissue atlas in an automated manner: we ID brain regions based on cell composition and quantify each cell type within each region 
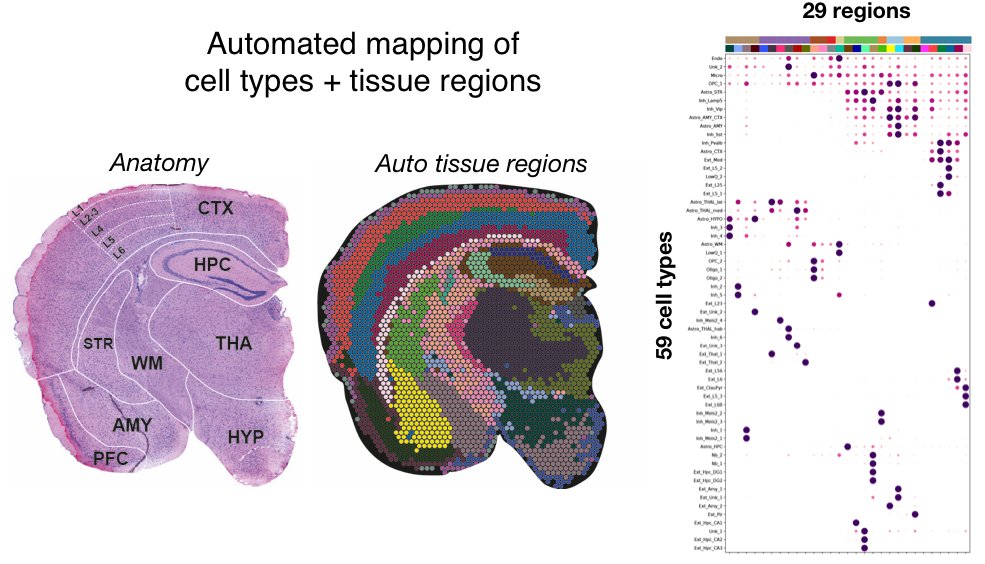
We also discover new biology: new regional astrocyte populations in the thalamus and hypothalamus! Most striking is a habenula-specific subtype - highly rare (0.1%) in our snRNAseq, closely related to other astrocytes, yet cell2location resolves it to a tiny brain region 

In the human lymph node, we can disentangle spatially interlaced immune cell states. We can identify spatially co-localised cell types by applying NMF to the cell2location output: beautifully identifies the cellular compartments of the tissue 

Finally, cell2location is versatile across technologies: we apply it to different single cell (10X, SmartSeq, cell/nuclei) and spatial methods (Visium and Slide-Seq) 

Check out cell2location on github.com/BayraktarLab/c…, see tutorials on our website cell2location.readthedocs.io. More updates to tutorials, including Google Colab notebook on the way.
Our amazing team & collaborators: @emmamarydann @artem_shmatko @AlexanderAivaz2 @hamish_king @LomakinAI @kedlian @immunobananna
@roserventotormo @MoritzGerstung
we built on @pymc_devs, Pyro, #scanpy Visium @10xGenomics And huge thanks @embl @emblebi @sangerinstitute
@roserventotormo @MoritzGerstung
we built on @pymc_devs, Pyro, #scanpy Visium @10xGenomics And huge thanks @embl @emblebi @sangerinstitute
• • •
Missing some Tweet in this thread? You can try to
force a refresh


