
Please check out our latest publication on pathological protein succinylation in humans:
"SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease"
doi.org/10.1038/s41467…
"SUCLA2 mutations cause global protein succinylation contributing to the pathomechanism of a hereditary mitochondrial disease"
doi.org/10.1038/s41467…
I first got involved in this project in 2016, and I gave an oral presentation on this at #USHUPO 2018.
Very fortunate to be part of this worldwide collaborative team, including: California, North Carolina, Wisconsin, Switzerland, and Sweden
Very fortunate to be part of this worldwide collaborative team, including: California, North Carolina, Wisconsin, Switzerland, and Sweden
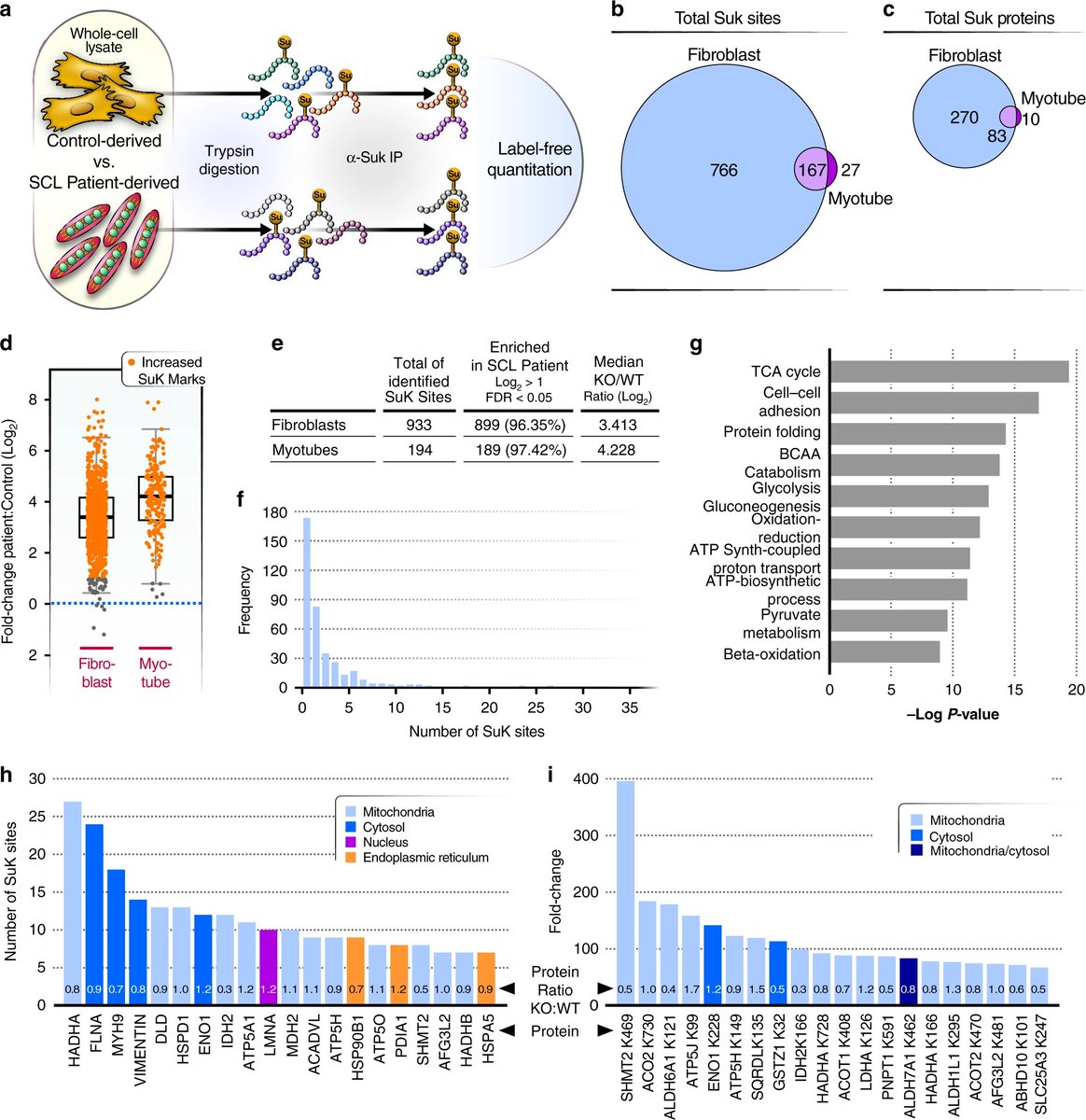
This started with Philipp's observation that succinyl-CoA binding proteins of the complex succinyl-CoA ligase (SCL) had higher succinylation than other proteins in the TCA cycle 

I normalized the published data to protein relative intensity and found that TCA cycle proteins (near the site of succinyl-CoA production) were more likely to be hypersuccinylated in SIRT5 KO than random chance 

We got fibroblasts and myotubes from patients with rare genetic succinyl-CoA ligase deficiency and found that, as expected, they had elevated succinyl-CoA 
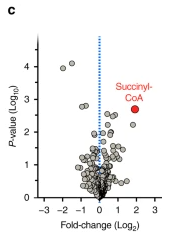
western blots suggested hyper succinylation in those patient cells, so I did succinyl proteomics analysis on those fibroblasts and myotubes and we found that almost every succinylation site was statistically higher in patient cells
this is also where you can see the magic of graphic designer help (Figure 2):
original figure graphic designer

original figure graphic designer

This is when fluency in R really helped: we wanted to compare the sites we found in human cells to those from Park et all SIRT5 KO in mouse embryonic fibroblasts. I wrote code to align the lysine residues between modified proteins from both human and mouse datasets
Figure 3 compares those datasets. We find that the fold change is much higher in cells from SCL patients versus SIRT5 KO mice. Apples to oranges in many ways but interesting:
original figure graphic designer

original figure graphic designer


Finally, having confirmed the hypersuccinylation was bad in this disease, Philipp made the same mutations in zebrafish, and also made a Sirt5 overexpresser to see if that would help rescue their phenotype despite disrupted TCA cycle 

Then they put the fish in a seahorse respirometer! The Sirt5 OE in the SUCLA2 knockout background had higher uncoupled respiration, and it lived longer than the SUCLA2 KO 

code for succinyl site analysis is here (sorry its messy) github.com/jessegmeyerlab…
This paper marks my temporary retirement from the field of protein acylation.
cliffhanger: I might come back
cliffhanger: I might come back
• • •
Missing some Tweet in this thread? You can try to
force a refresh


