
Excited to share first preprint from my postdoc work in Stevens lab: “Single Cell Sequencing Reveals Glial Specific Responses to Tissue Processing & Enzymatic Dissociation in Mice and Humans” a thread below #singlecell #microglia #methodsmatter 1/n
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
What started as small internal pilot in the lab, turned into full-fledged paper. I’ll detail some of the key findings in thread below. But first want to take a second and thank all of our fantastic colleagues and collaborators without whom this project wouldn’t be possible 2/n
Help from many fantastic members of Stevens lab @thammondglia , Lasse, Alec, @ConnorDufort , Sarah, @soyonhonglab and of course my mentor @stevenslab1. Continuing fantastic collaboration and advice from @macosko and his lab Tushar, Chuck, @Abdul_Squared , Naeem, and Velina. 3/n
Adam Young an amazing neurosurgeon in the UK and his collaborators who helped with the fresh neurosurgical tissue. Also of course @10XGenomics for making such a fantastic method widely available and inviting me to talk about this work last month. 4/n
FYI This will be very long one so some tl;dr up front. METHODS MATTER! The most basic conclusion is: scRNA-seq is incredibly powerful, but its power is predicated on being able to faithfully measure cell-type-specific expression patterns ex vivo w/o altering those patterns 5/n
Also didn’t want to stick this at the end but since starting single cell work at beginning of my postdoc I’ve seen just how amazing this field can be especially in regards to sharing data and reproducibility and it’s something I strongly believe in. 6/n
Nearly all raw data is deposited & freely available (1 controlled access deposit is in progress). All of the code to reproduce the objects used for analysis can be found at GitHub repo here: github.com/samuel-marsh/M…
ReadME inspired by @rmassonix’s on similar topic in PBMCs. 7/n
ReadME inspired by @rmassonix’s on similar topic in PBMCs. 7/n
Ok now let’s dig in! I work on microglia and we know microglia, like all tissue-resident macrophages are extremely sensitive to their environment. Our previous studies optimized cold mechanical dissociation method to isolate microglia for scRNA-seq 8/n
However, many other popular microglia isolation protocols utilize enzymatic digestion when working with brain tissue. We suspected this would induce ex vivo changes in microglia prior to sequencing. 9/n
Therefore, we also optimized an inhibitor protocol to block transcription and translation during the isolation process to prevent any ex vivo changes. This was the design of our first experiment studying sorted microglia 10/n 
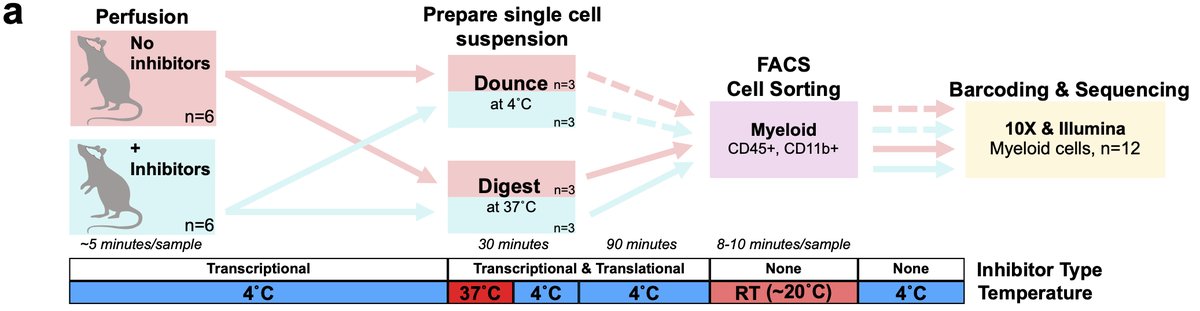
In addition to populations we’d previously characterized, we found a population we refer to as “exAM”, nearly exclusive to samples digested w/enzymes and no inhibitors. We were happy to confirm that our previous cold Dounce protocol was free of artifacts even w/o inhibitors. 11/n 
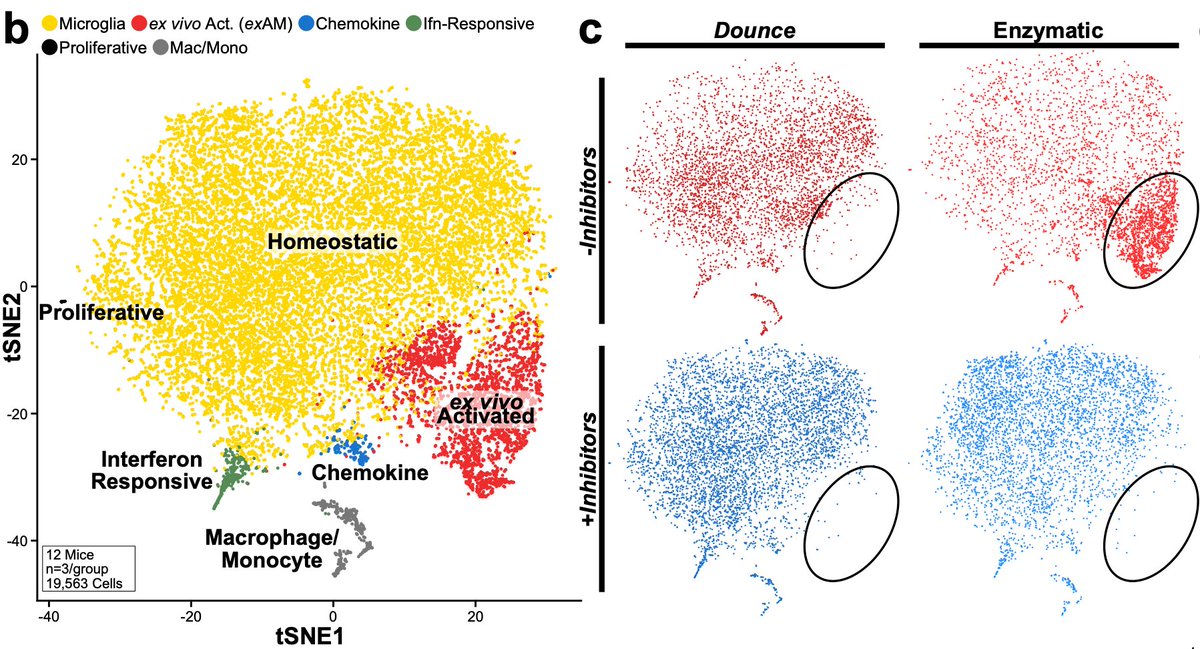
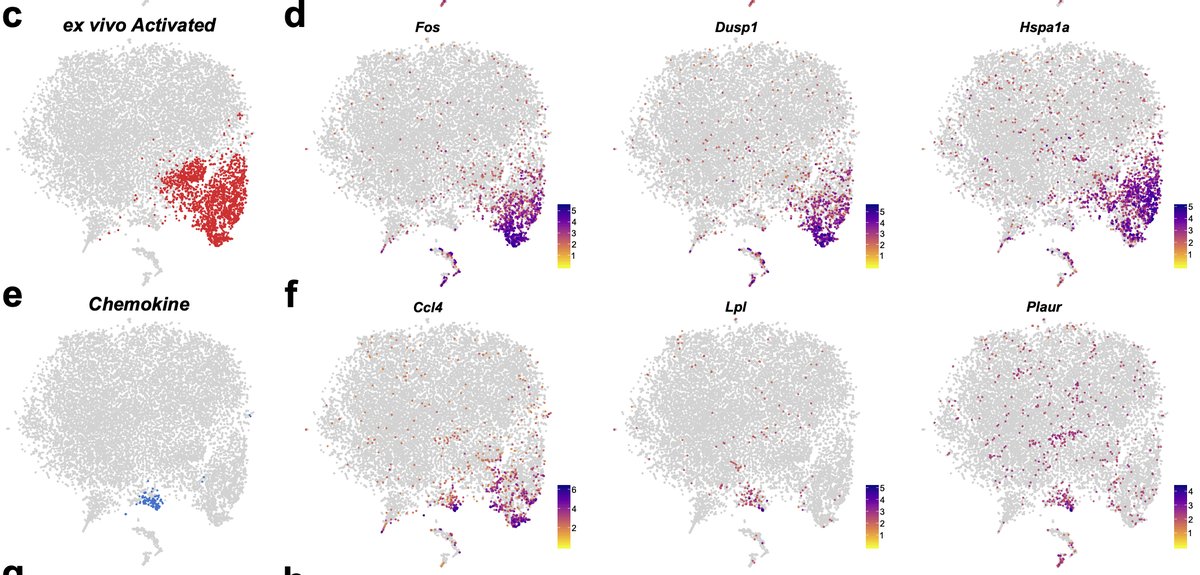
This cluster was characterized by expression of IEGs, stress-related genes, and inflammatory genes. Unfortunately, it does overlap with a rare “true” population of chemokine secreting microglia across some markers which does complicate experiments where enzymes were used. 12/n 
Given how prevalent this signature was in our data we performed reanalysis of several literature datasets across enzymes and seq technologies. All datasets exhibited significant ex vivo activation in microglia/myeloid pops except our previous cold Dounce study 13/n 
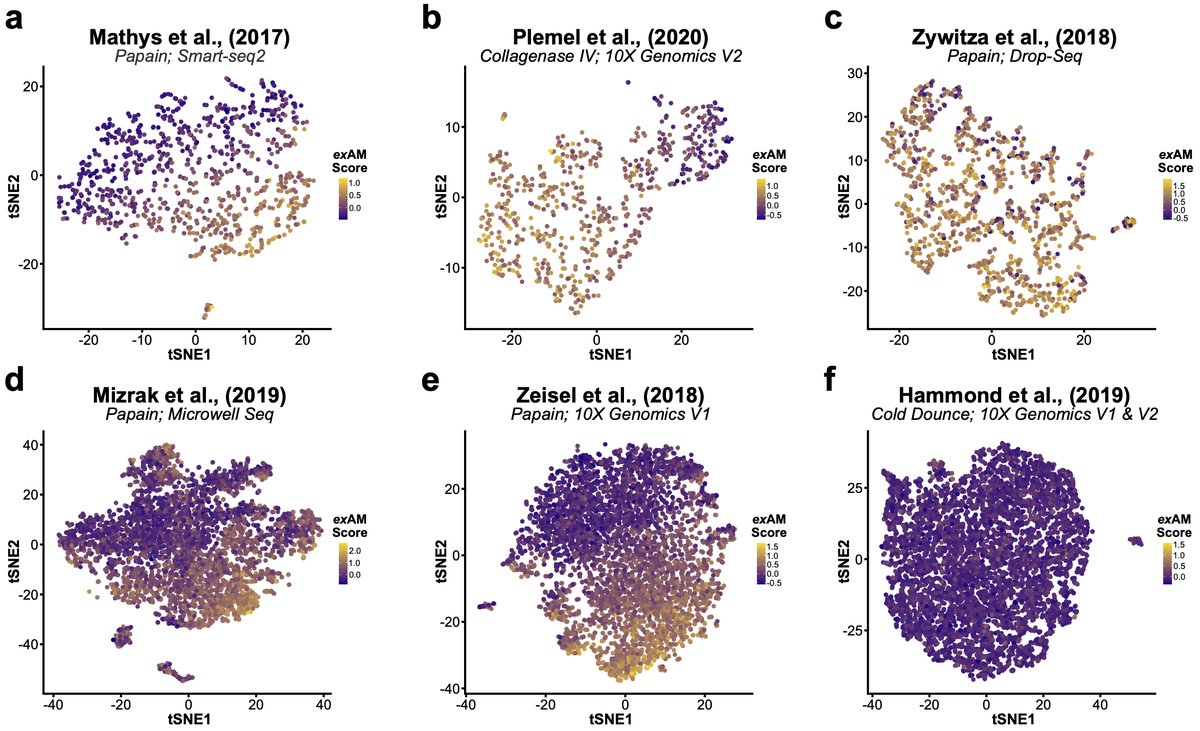
This means we need to look at previous microglial studies w/enzymatic digestion (or room temp. steps) with a more critical eye and potentially re-evaluate some of their conclusions. Microglia/myeloid cells are of particular importance here, but more on that in bit. 14/n
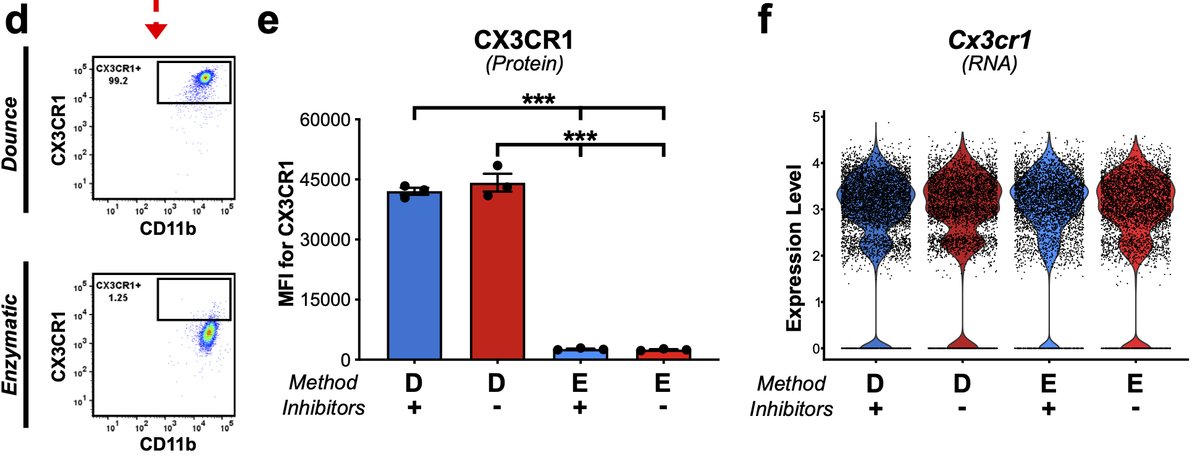
One other important aside here that is crtical for CyTOF or CITE-Seq-based approaches. Enzymes cleave some cell surface proteins & different enzymes cleave differently. This has been previously been shown via flow in peripheral immunology 15/n
Microglia digested with papain exhibited nearly 20-fold decrease in MFI compared to Dounce samples. This is critical consideration for any experiment measuring surface proteins, comparing digested to undigested tissues, or comparing tissues digested with different enzymes 15/n 
The other aside is something I feel like I shouldn’t have to say but REPLCIATES MATTER! It doesn’t matter if you are comparing 100,000 cells from one sample to 100,000 from another, without true biological replicates you can’t accurately assess those samples 16/n
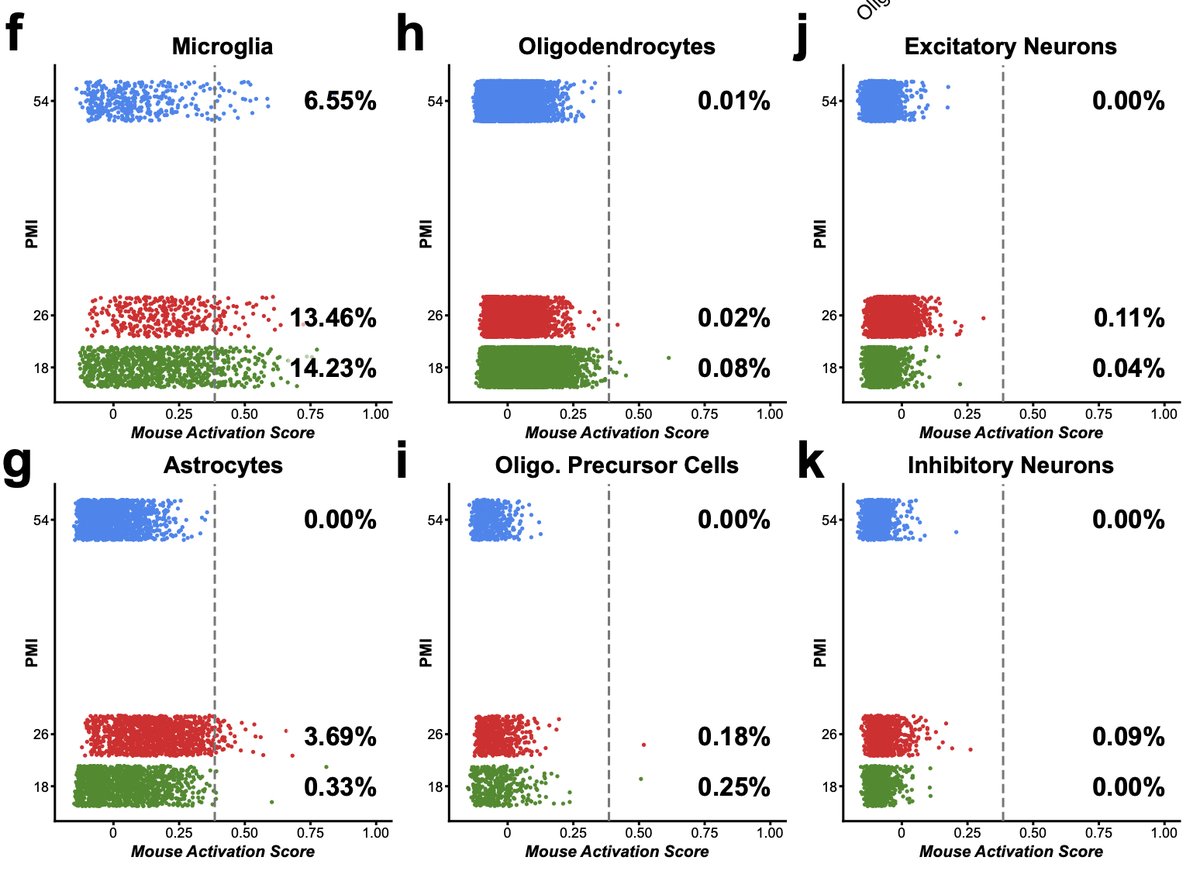
The same is true if you pool samples without ability to demultiplex post-10X. Here is example from separate microglia experiment. 4 samples processed with cold Dounce. Overall activation score is quite low, except Sample 4 where the number of cells increases 10 fold! 17/n 
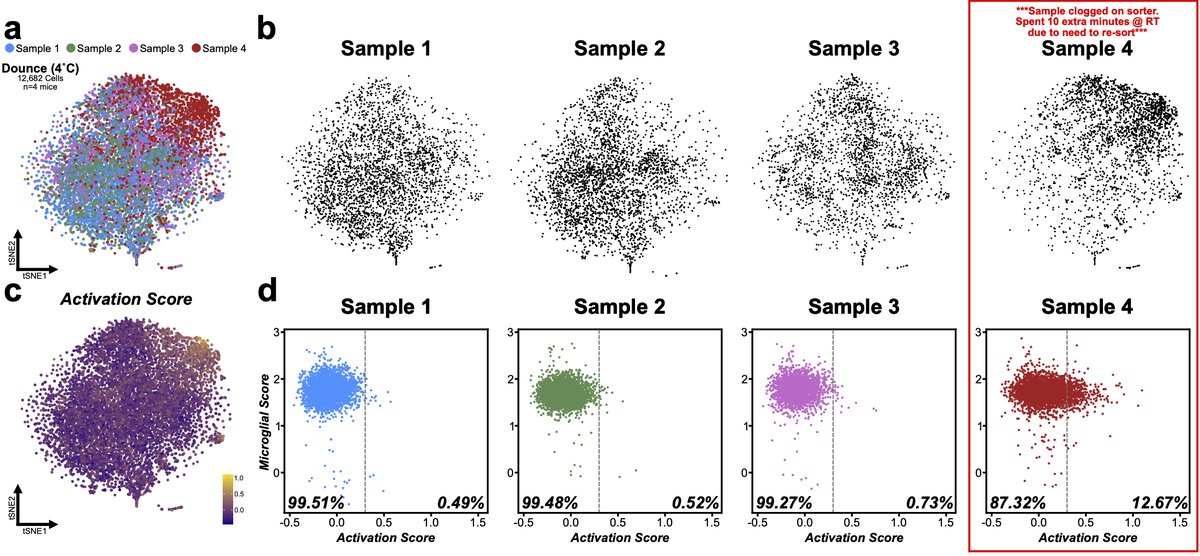
In this case we know part of the story. Sample clogged on sorter and had to be resorted so it spent extra time at RT and had extra set of temp changes before resort. Which shows even minimal issues can have big impact. 18/n
Without replicates if I compared S1 to S4 I might attribute a biological change to a what is actually a technical artifact. Unfortunately, this has become too common in scRNA pubs, even some methods papers but it’s time field & editors stopped allowing these things through. 19/n
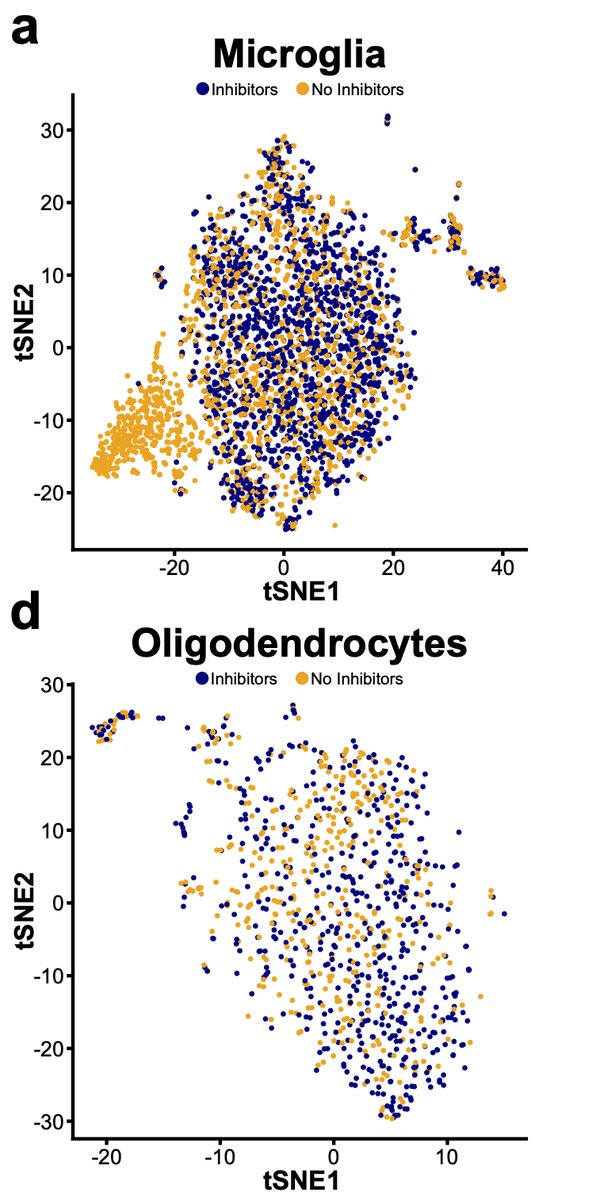
Next, we wondered whether other non-microglial CNS cells might also exhibit changes following isolation so we performed similar experiment profiling all CNS cells. We find that microglia are preferentially sensitive to this aberrant profile 20/n 


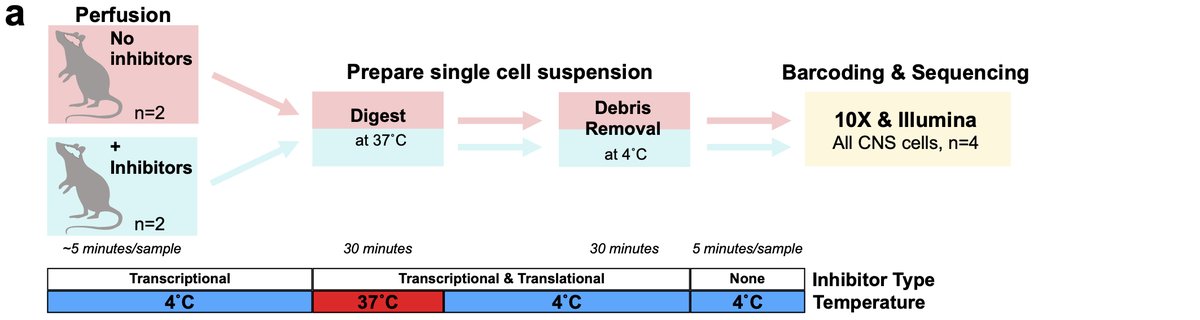
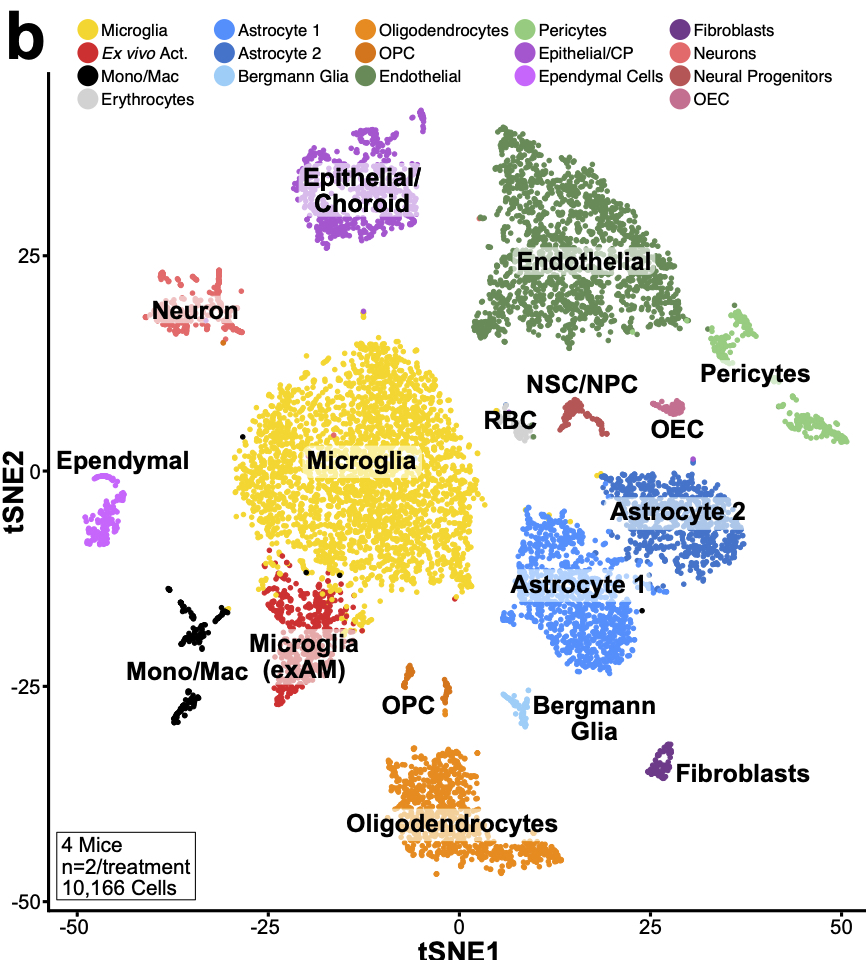
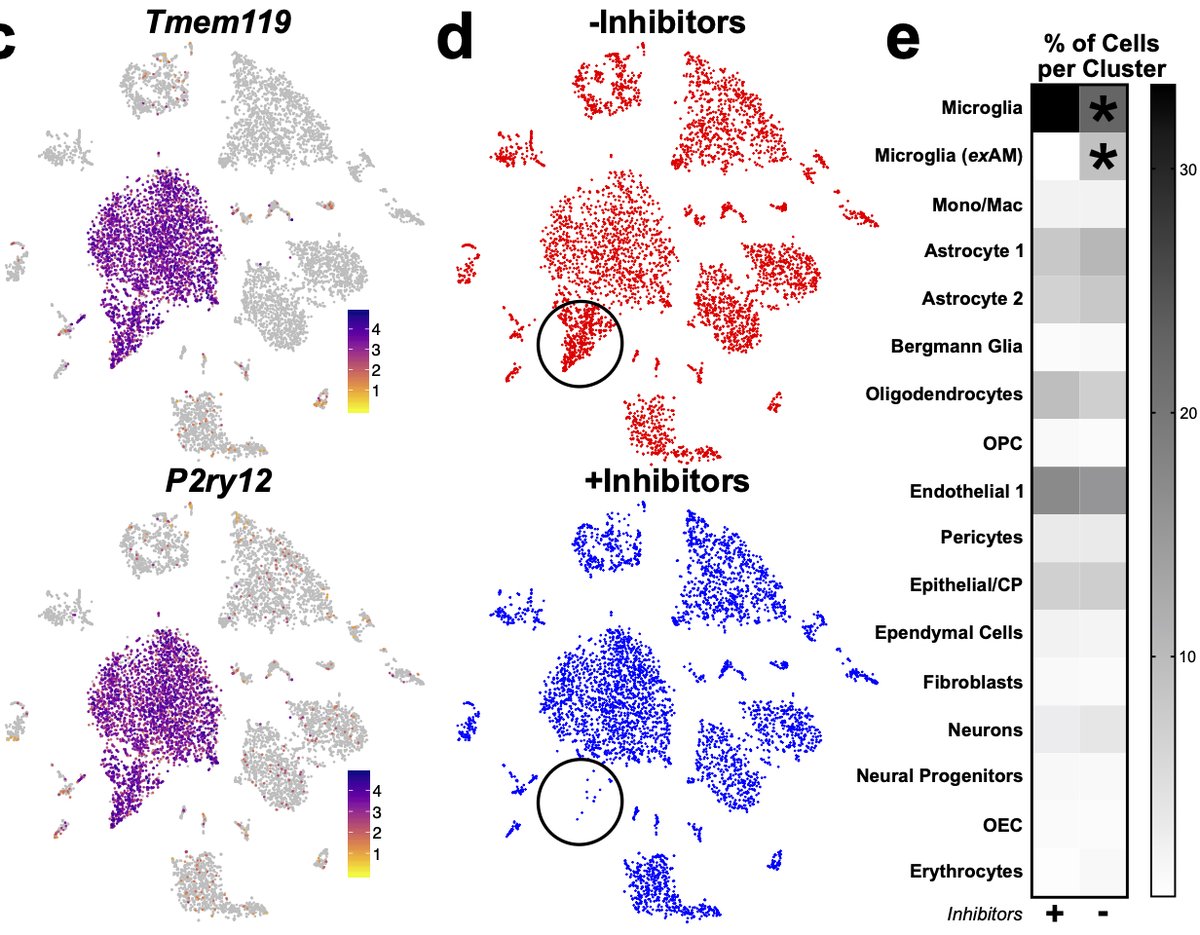
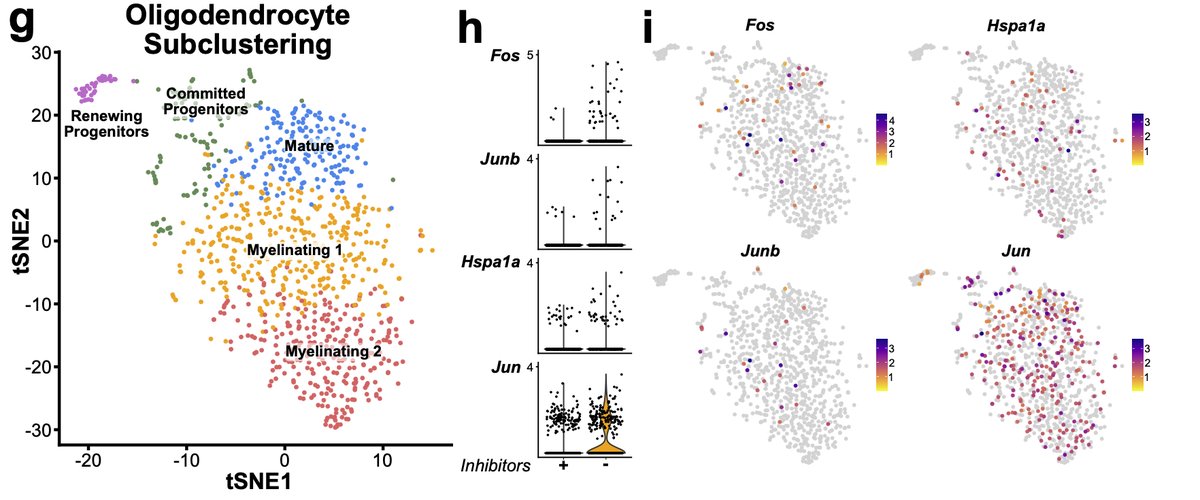
However, while not significant if we look deeper at other cell types some changes are starting to occur. For instance, looking at oligodendrocyte lineages, while not significant, we see the start of some induction of genes in the signature. 21/n 

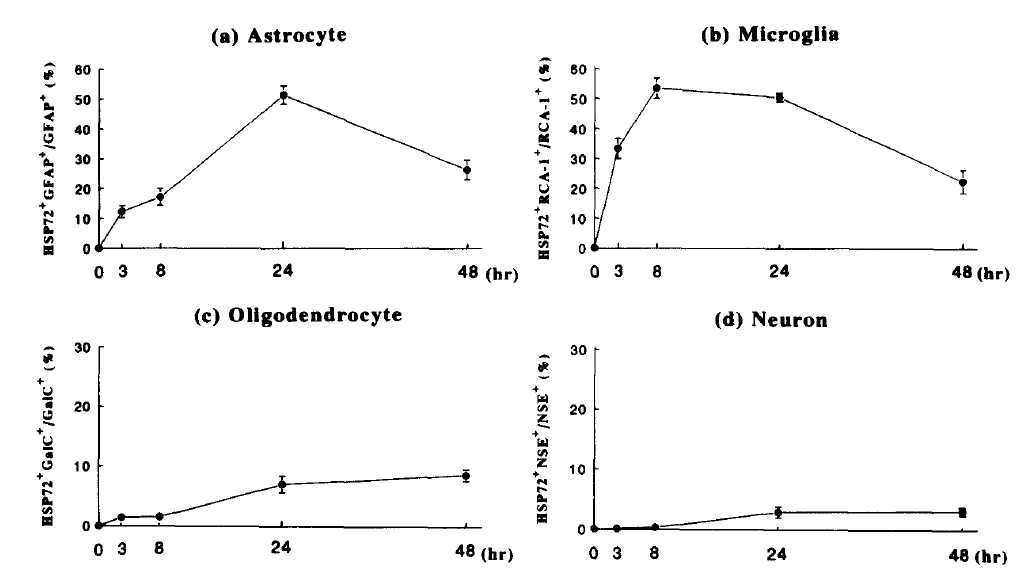
This fits with previous literature on the speed and magnitude of cell response to stress. For instance, in primary human cells Satoh & Kim show that microglia and astrocytes respond in equal magnitude but on completely different time scales 22/n pubmed.ncbi.nlm.nih.gov/7982058/ 
Again, our inhibitor protocol stopped ex vivo activation in microglia. The advantage of our solution is that it leverages the ability to use existing, often highly-optimized dissociation protocols with simple addition, which saves issues with creating entirely new protocols. 23/n
For instance, while cold-active proteases have worked for other tissues, we have struggled to adapt them to CNS for scRNA applications. This has mainly been hampered by lack of cold-active specific DNase. Even DNase from arctic shrimp has optimal function at 37 degrees… 24/n
We next wondered whether similar artifactual signature might be present in human post-mortem (PM) tissue. We performed snRNA-seq of >47,000 nuclei (3 donors) with wide spread of PMIs. Our original hypothesis was: PMI would be correlated with the presence of this signature. 25/n 
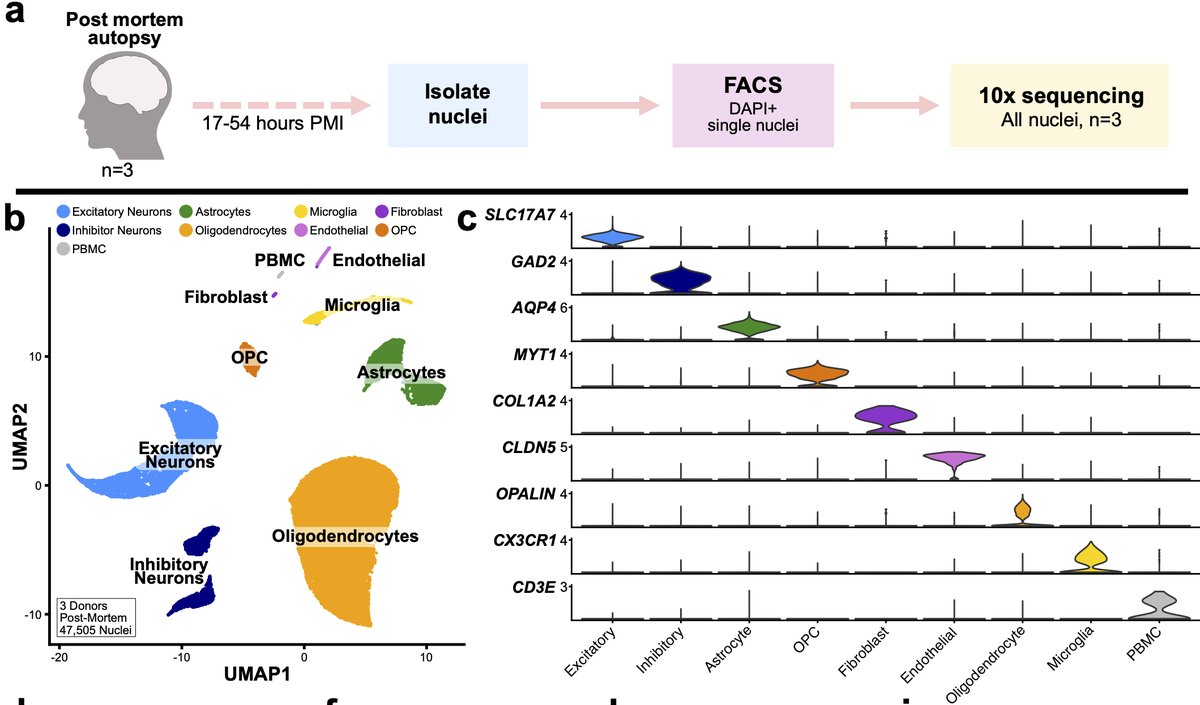
When we performed module scoring using the mouse signature, we found that microglia and some astrocytes had scores above threshold but that the percentage of cells above threshold per sample did not track with PMI 26/n 
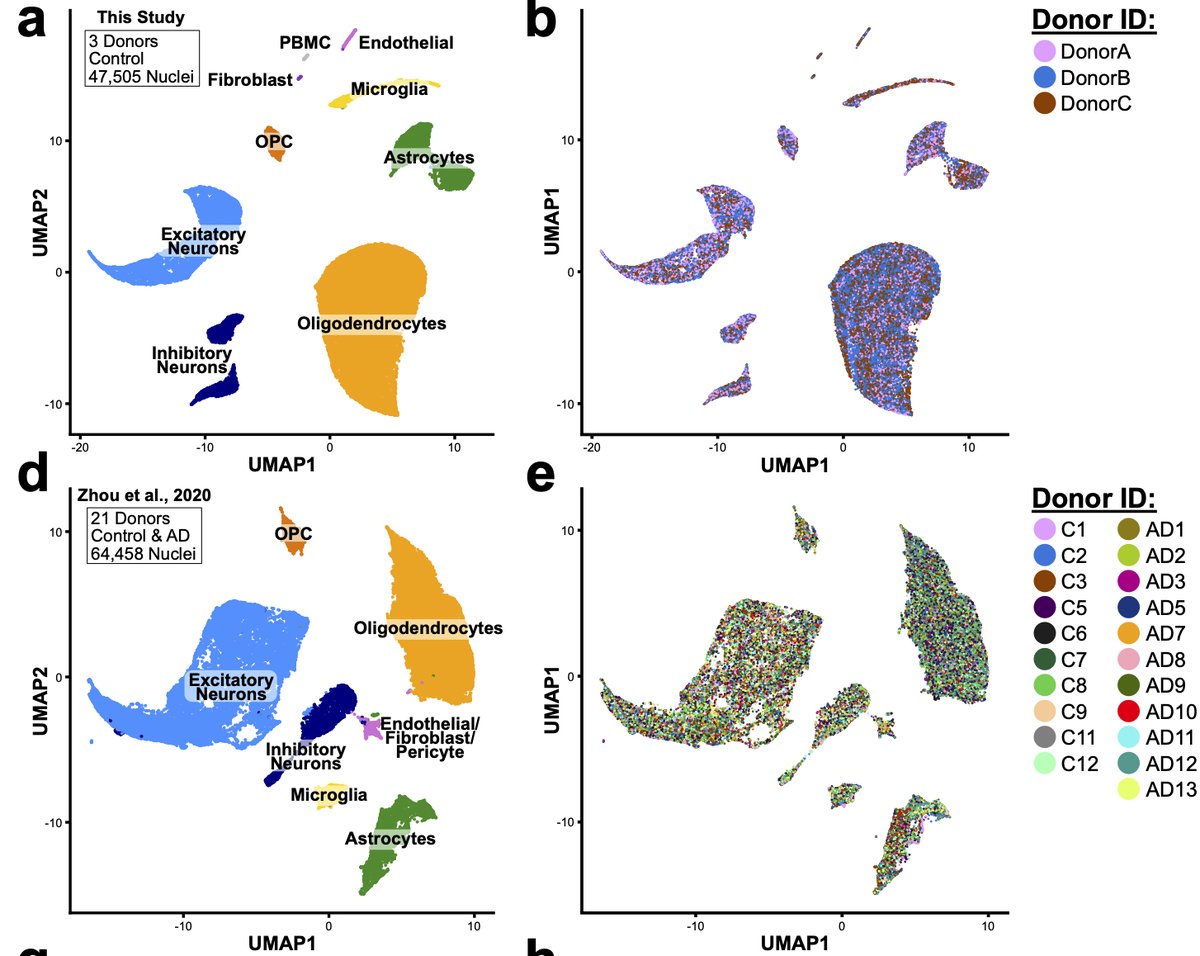
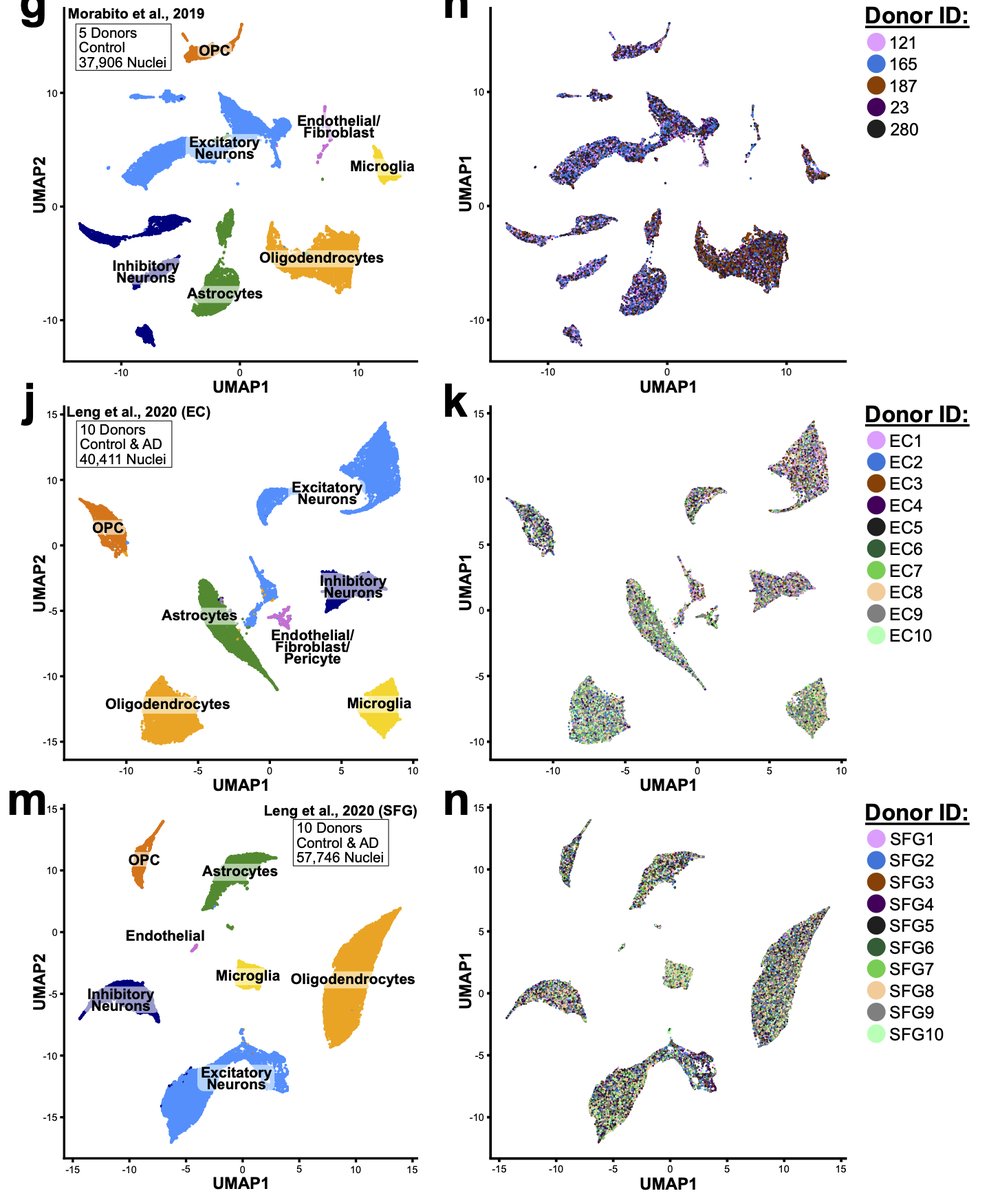
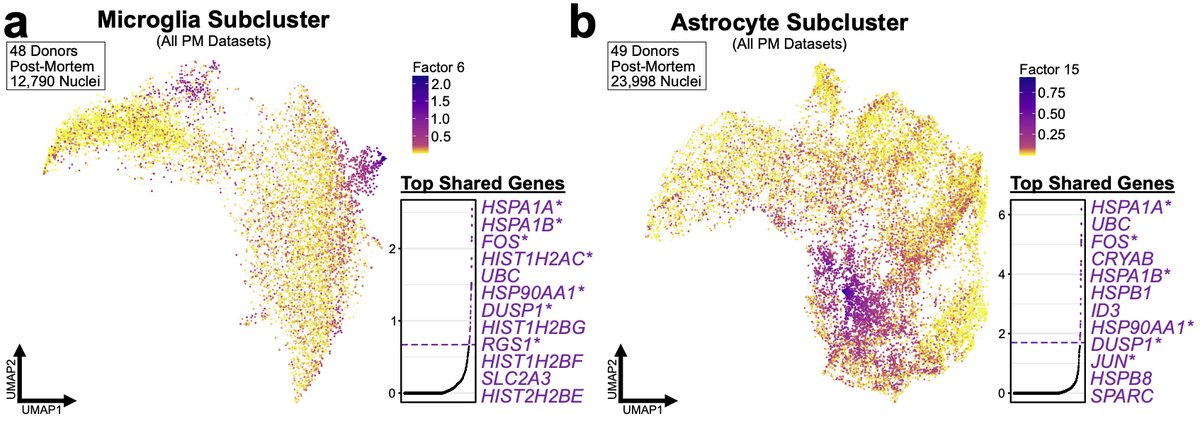
To power our analysis, we performed reanalysis of 4 literature datasets for total of 49 donors and nearly 250K total nuclei. We found that proportions of microglia and astrocytes were enriched for this artificial signature across all of the datasets. 27/n 

However, we now wondered whether we could detect this signature without a priori knowledge of what genes might be in it. For all of our human analyses we used the integrative method LIGER developed by @macosko and @labwelch that utilizes iNMF-method to integrate datasets. 28/n
We used the shared factors generated during LIGER analysis to look for similar signatures. We found LIGER factors in both PM microglia and astrocytes that were similar to the artificial mouse signature. However, as we examined PM data we couldn’t “confirm” it was artificial. 29/n 
To get at this we collaborated with Dr. Adam Young and team to collect freshly-resected neurosurgical tissue. We snap-froze half the tissue and incubated the other half for 6 hours in aCSF to create a technical variable of time-delay before freezing. 30/n 
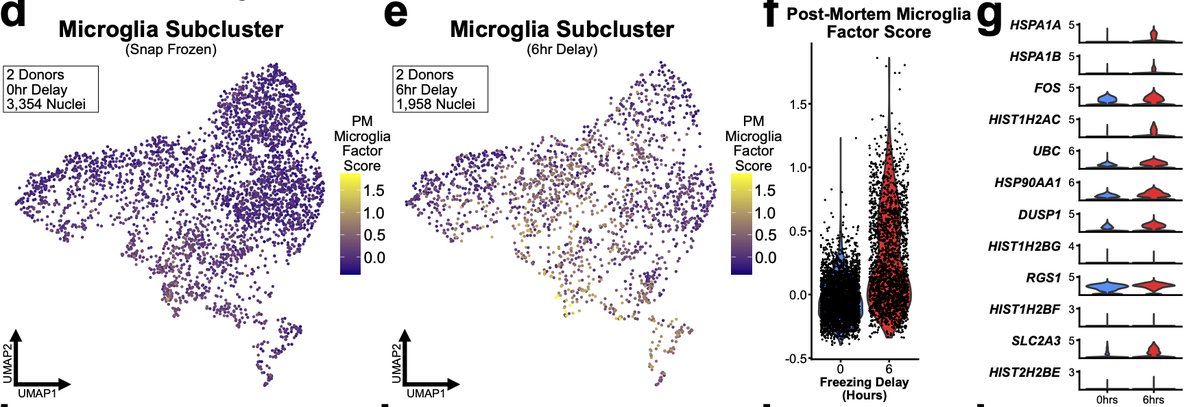
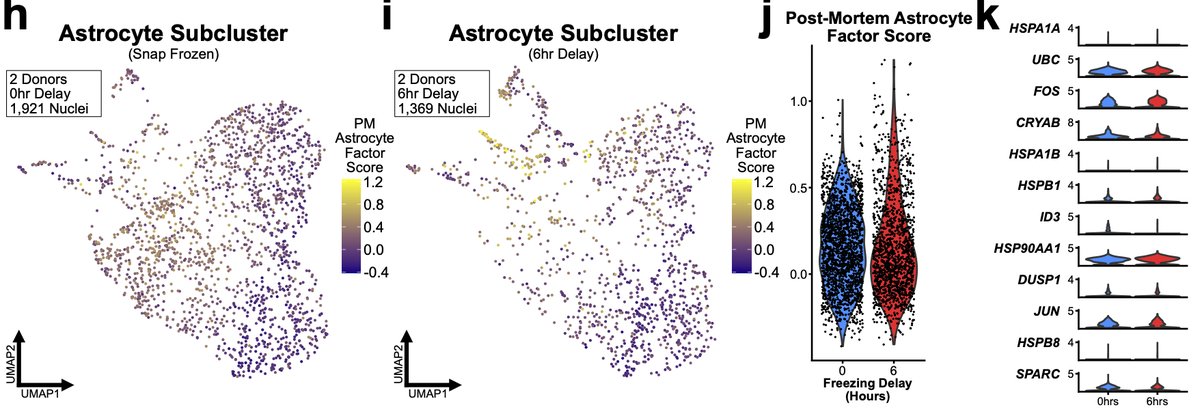
We found that microglia from 6hr samples significantly upregulated of nearly all genes in the microglial PM LIGER factor. But 6hr astrocytes did not exhibit similar significant changes. We believe this again is due to the different speeds at which different cells respond. 31/n 

Finally, when we quantified the percent of nuclei above threshold for either PM microglial or astrocyte factors vs. meta-data variables and PMI was not correlated but age of the donor was. 32/n 

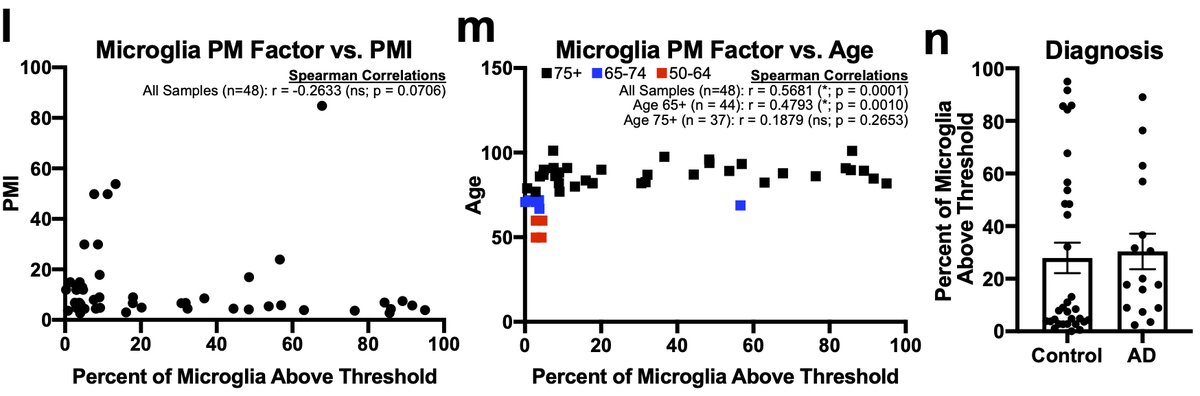
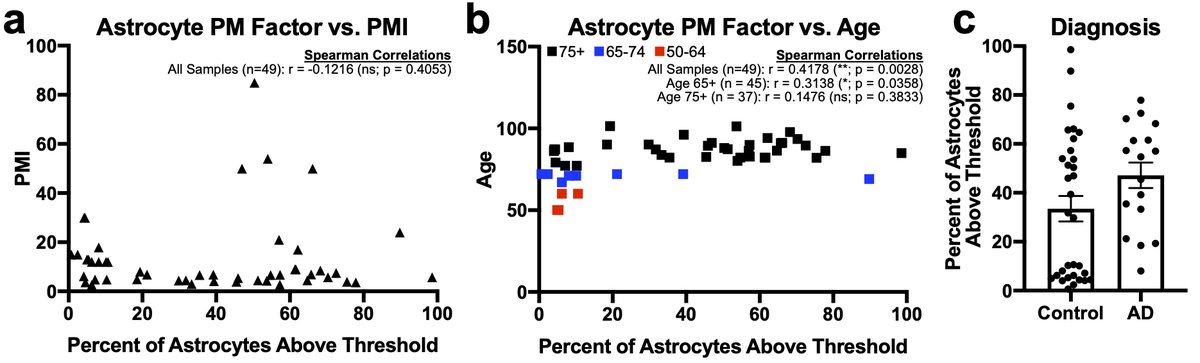
However, the small number of younger samples all came from one dataset and if we excluded those younger than 75 the correlation went away. Given the between donor variability in the 75+ cohort we believe that more studies of middle-aged 33/n
…patients are needed before we can conclude anything regarding age. Lastly, there was no difference between AD and control subjects. One final aside continuing the #methodsmatter theme is that protocol and sequencing tech make huge difference for nuclei 34/n
These are the median genes and UMIs/nucleus per sample across all the studies and differences for microglia are even more pronounced!! Nearly 1000! median genes/nucleus difference on average between our dataset and the next highest dataset. 35/n 

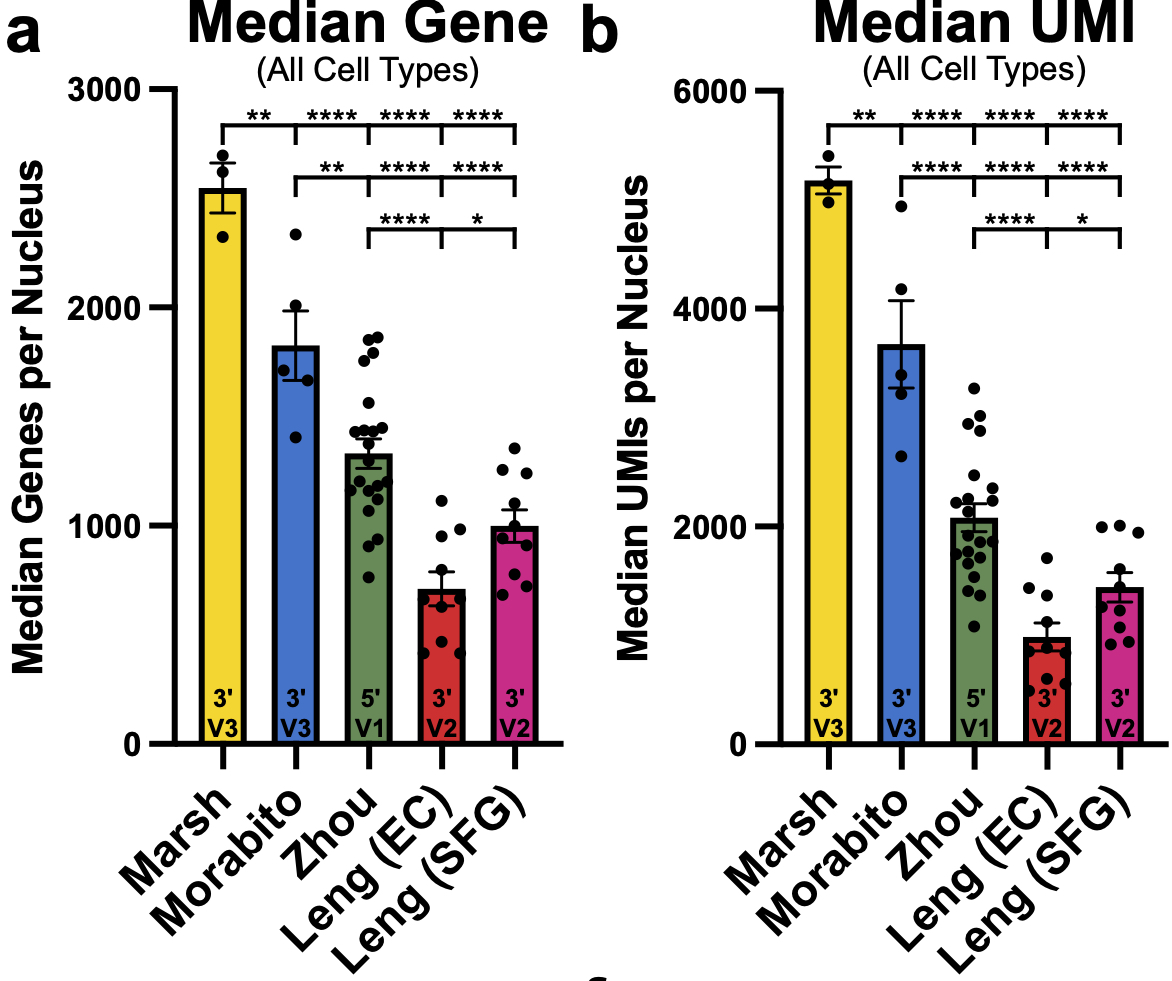
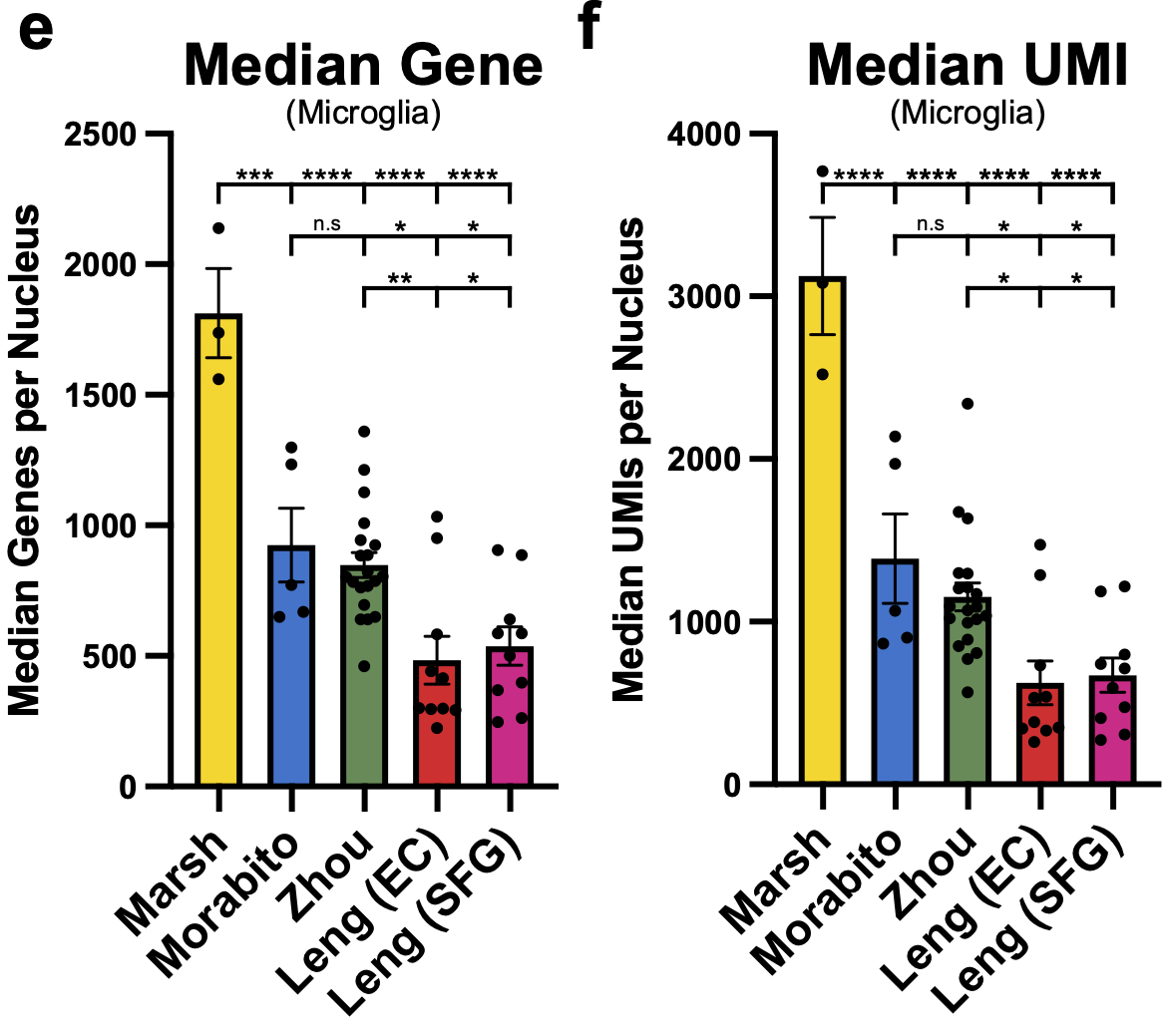
Hopefully as more and more post-mortem datasets with newer kits become available so too will our ability to resolve this post-mortem signature and the factors behind it (also please publish as much meta data as you are able! For this and other general analytical reasons) 36/n
I think that just about wraps up my longest ever twitter thread. But check out the preprint for more info and if you have any questions or want to discuss the results or implementing our protocol please reach out and we are happy to help! 37/37
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh


